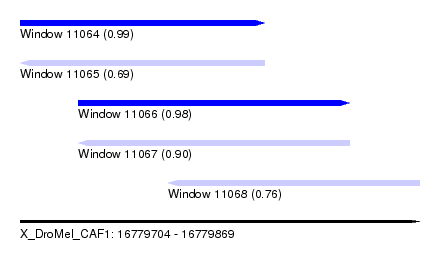
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,779,704 – 16,779,869 |
| Length | 165 |
| Max. P | 0.986141 |

| Location | 16,779,704 – 16,779,805 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -17.53 |
| Consensus MFE | -16.78 |
| Energy contribution | -16.56 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16779704 101 + 22224390 UUAGCAAAAUAACAAUGCAACGAAAGAAGAAAAGCAUAAAAACGCAAGGGGAGAACGCAAGUGAUUCAUGCGAAUAAAUAAAUGACUAUCCAAUUGCAAAU ...............((((((....).......((((.....(....).(((..((....))..)))))))......................)))))... ( -17.60) >DroSec_CAF1 16601 101 + 1 UUAGCAAAAUAACAAUGCAACAAAAGAAGAAAUGCAUAAAAUCGCAAGGGGAGAACGCAAGUGAUUCAUGCGAAUAAAUAAAUGACUAUCCAAUUGCAAAU ...((((.......(((((.............)))))....(((((...(((..((....))..))).)))))....................)))).... ( -19.72) >DroYak_CAF1 16483 97 + 1 UUAGCAAAAUAACAAUGCAACAAAAGAAGAAAAGCAUAAAAACGUAAAGGGAGAACGCAAGUGAUUCAUGCGAAUA----AAUGACUAUCCAAUUGCAAAU ...((((.......((((...............)))).....((((...(((..((....))..))).))))....----.............)))).... ( -15.26) >consensus UUAGCAAAAUAACAAUGCAACAAAAGAAGAAAAGCAUAAAAACGCAAGGGGAGAACGCAAGUGAUUCAUGCGAAUAAAUAAAUGACUAUCCAAUUGCAAAU ...((((.......((((...............)))).....((((...(((..((....))..))).)))).....................)))).... (-16.78 = -16.56 + -0.22)


| Location | 16,779,704 – 16,779,805 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -14.99 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16779704 101 - 22224390 AUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCACUUGCGUUCUCCCCUUGCGUUUUUAUGCUUUUCUUCUUUCGUUGCAUUGUUAUUUUGCUAA ...((((((.((((((........))))))(((((((.....((((.........))))..)))))))............))))))............... ( -17.00) >DroSec_CAF1 16601 101 - 1 AUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCACUUGCGUUCUCCCCUUGCGAUUUUAUGCAUUUCUUCUUUUGUUGCAUUGUUAUUUUGCUAA ...((((((.((((((........))))))(((((((....(((((.........))))).)))))))............))))))............... ( -18.40) >DroYak_CAF1 16483 97 - 1 AUUUGCAAUUGGAUAGUCAUU----UAUUCGCAUGAAUCACUUGCGUUCUCCCUUUACGUUUUUAUGCUUUUCUUCUUUUGUUGCAUUGUUAUUUUGCUAA ...((((((.((((((....)----)))))(((((((......((((.........)))).)))))))............))))))............... ( -16.50) >consensus AUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCACUUGCGUUCUCCCCUUGCGUUUUUAUGCUUUUCUUCUUUUGUUGCAUUGUUAUUUUGCUAA ...((((((.((((((........))))))(((((((......((((.........)))).)))))))............))))))............... (-14.99 = -15.43 + 0.45)


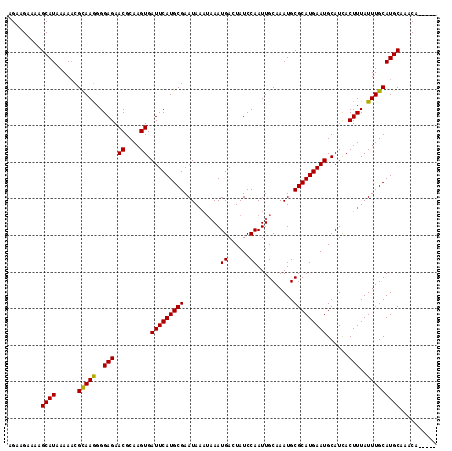
| Location | 16,779,728 – 16,779,840 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.46 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -24.06 |
| Energy contribution | -23.62 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16779728 112 + 22224390 AGAAGAAAAGCAUAAAAACGCAAGGGGAGAACGCAAGUGAUUCAUGCGAAUAAAUAAAUGACUAUCCAAUUGCAAAUGCGCAUGAAUGCAUCACUUUAUUUGCAUGCAAACA----- .........((((......(((((..(((.((....)).(((((((((......(((.((......)).)))......)))))))))......)))..))))))))).....----- ( -25.00) >DroSec_CAF1 16625 112 + 1 AGAAGAAAUGCAUAAAAUCGCAAGGGGAGAACGCAAGUGAUUCAUGCGAAUAAAUAAAUGACUAUCCAAUUGCAAAUGCGCAUGAAUGCAUCACUUUAUUUGCAUGCAAUCA----- ........(((((....(((((...(((..((....))..))).))))).(((((((((((..........((....))(((....))).))).)))))))).)))))....----- ( -26.20) >DroYak_CAF1 16507 113 + 1 AGAAGAAAAGCAUAAAAACGUAAAGGGAGAACGCAAGUGAUUCAUGCGAAUA----AAUGACUAUCCAAUUGCAAAUGCGCAUGAAUGCAUUACUUUAUUUGCAUGCAAGCCUUUAU ...................(((((((((..((....))..))((((((((((----((.............((....))(((....))).....))))))))))))....))))))) ( -25.30) >consensus AGAAGAAAAGCAUAAAAACGCAAGGGGAGAACGCAAGUGAUUCAUGCGAAUAAAUAAAUGACUAUCCAAUUGCAAAUGCGCAUGAAUGCAUCACUUUAUUUGCAUGCAAACA_____ .........((((......(((((..(((.((....)).(((((((((..........((......))..........)))))))))......)))..))))))))).......... (-24.06 = -23.62 + -0.44)

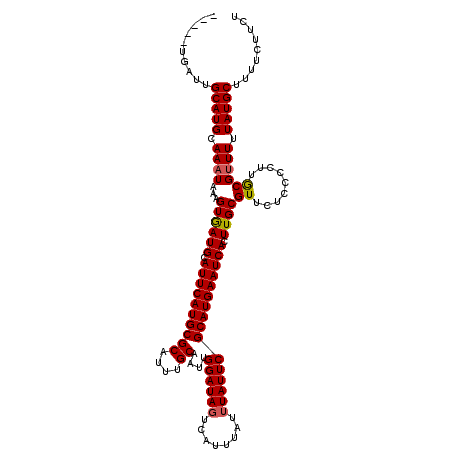
| Location | 16,779,728 – 16,779,840 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.46 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -22.41 |
| Energy contribution | -22.63 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16779728 112 - 22224390 -----UGUUUGCAUGCAAAUAAAGUGAUGCAUUCAUGCGCAUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCACUUGCGUUCUCCCCUUGCGUUUUUAUGCUUUUCUUCU -----.....(((((.((((.(((((((....((((((((....))....((((((........)))))))))))))))))))(((.........))))))).)))))......... ( -27.20) >DroSec_CAF1 16625 112 - 1 -----UGAUUGCAUGCAAAUAAAGUGAUGCAUUCAUGCGCAUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCACUUGCGUUCUCCCCUUGCGAUUUUAUGCAUUUCUUCU -----.((.((((((.((((.(((((((....((((((((....))....((((((........)))))))))))))))))))(((.........))).))))))))))..)).... ( -28.00) >DroYak_CAF1 16507 113 - 1 AUAAAGGCUUGCAUGCAAAUAAAGUAAUGCAUUCAUGCGCAUUUGCAAUUGGAUAGUCAUU----UAUUCGCAUGAAUCACUUGCGUUCUCCCUUUACGUUUUUAUGCUUUUCUUCU ...((((...(((((.((((((((.(((((((((((((((....))....((((((....)----)))))))))))).....))))))....))))..)))).)))))...)))).. ( -25.80) >consensus _____UGAUUGCAUGCAAAUAAAGUGAUGCAUUCAUGCGCAUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCACUUGCGUUCUCCCCUUGCGUUUUUAUGCUUUUCUUCU ..........(((((.((((...((((((.((((((((((....))....((((((........)))))))))))))))).))))((.........)))))).)))))......... (-22.41 = -22.63 + 0.23)


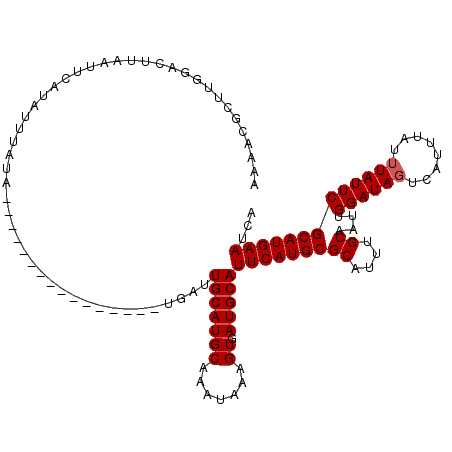
| Location | 16,779,765 – 16,779,869 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -17.80 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16779765 104 - 22224390 AAAACGCUUGGACUUAAUUCAUAUUUAUA----------------UGUUUGCAUGCAAAUAAAGUGAUGCAUUCAUGCGCAUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCA ....((((((((.....))).(((((.((----------------((....)))).)))))))))).((.((((((((((....))....((((((........)))))))))))))))) ( -22.90) >DroSec_CAF1 16662 104 - 1 AAAACGCUUGUACUUAAUUCAUAUUUAUA----------------UGAUUGCAUGCAAAUAAAGUGAUGCAUUCAUGCGCAUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCA .....((...(((((...(((((....))----------------)))(((....)))...)))))..))((((((((((....))....((((((........)))))))))))))).. ( -23.60) >DroYak_CAF1 16544 116 - 1 AAAACGGAUGGAUUUAAUACACAUUUAAACAUUUAAACAUAUAAAGGCUUGCAUGCAAAUAAAGUAAUGCAUUCAUGCGCAUUUGCAAUUGGAUAGUCAUU----UAUUCGCAUGAAUCA .....(((((...((((.......)))).)))))...............(((((((.......)).)))))(((((((((....))....((((((....)----))))))))))))... ( -20.20) >consensus AAAACGCUUGGACUUAAUUCAUAUUUAUA________________UGAUUGCAUGCAAAUAAAGUGAUGCAUUCAUGCGCAUUUGCAAUUGGAUAGUCAUUUAUUUAUUCGCAUGAAUCA .................................................(((((((.......)).)))))(((((((((....))....((((((........)))))))))))))... (-17.80 = -18.13 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:15 2006