
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,776,785 – 16,776,902 |
| Length | 117 |
| Max. P | 0.999803 |

| Location | 16,776,785 – 16,776,902 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
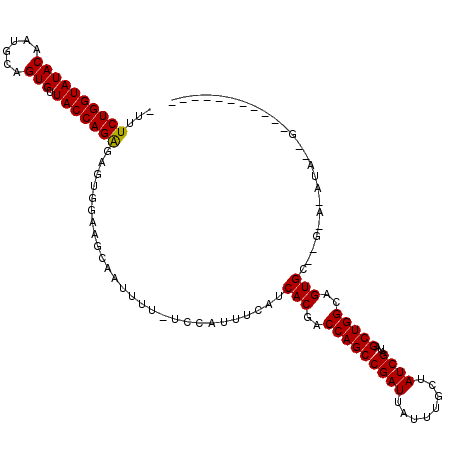
>X_DroMel_CAF1 16776785 117 + 22224390 -UUUCUGGUAUACAAUGCAGUGUUACCAGAGCGUGGAAGUAAUUUU-UACAUUUAAUCACGACCAGCCGAUUAUUUGCUAUCGAUAGCUGGCAGUGCG-GUAUAUAUCGUUUUUUAAAUA -.(((((((((((......))).)))))))).(((((((...))))-)))((((((.(((..(((((((((........))))...)))))..)))((-((....))))....)))))). ( -29.70) >DroSec_CAF1 13819 96 + 1 -UUUCUGGUAUACAAUGCAGUGUUACCAGAGAGUGGAAGCAAUUUUAUCCAUUUCAUCACGACCAGCCGAUUACUUGCUAUCGAUAGCUGGCAGUGC----------------------- -..((((((((((......))).)))))))(((((((..........)))))))...(((..(((((((((........))))...)))))..))).----------------------- ( -29.60) >DroYak_CAF1 13818 109 + 1 UUUUCUGGUAUACAAUGCAGUGUUACCAGGUUGUGAAAACAUUUUU-UCUUAUUCAUCACGACCAGCCGAUUAUUUUCUAUCGAUAGCUGGCGGUGCCGGAAAAUACUGA---------- (((((((((((((......)))).(((.((((((((..........-.........)))))))).((((.(((((.......))))).))))))))))))))))......---------- ( -30.81) >consensus _UUUCUGGUAUACAAUGCAGUGUUACCAGAGAGUGGAAGCAAUUUU_UCCAUUUCAUCACGACCAGCCGAUUAUUUGCUAUCGAUAGCUGGCAGUGC__G_A_AUA__G___________ ...((((((((((......))).)))))))...........................(((..(((((((((........))))...)))))..)))........................ (-24.22 = -24.00 + -0.22)


| Location | 16,776,785 – 16,776,902 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -24.07 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.12 |
| SVM RNA-class probability | 0.999803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
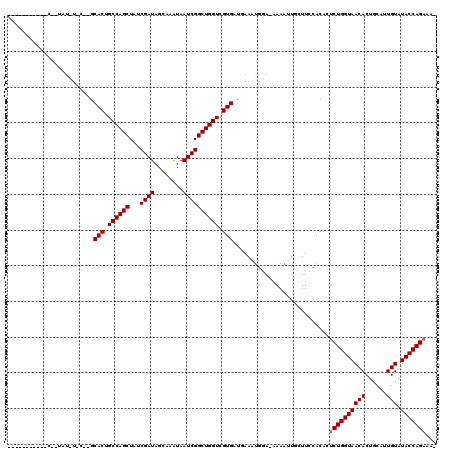
>X_DroMel_CAF1 16776785 117 - 22224390 UAUUUAAAAAACGAUAUAUAC-CGCACUGCCAGCUAUCGAUAGCAAAUAAUCGGCUGGUCGUGAUUAAAUGUA-AAAAUUACUUCCACGCUCUGGUAACACUGCAUUGUAUACCAGAAA- ..............(((((..-..(((.((((((...((((........)))))))))).))).....)))))-................((((((((((......))).)))))))..- ( -27.10) >DroSec_CAF1 13819 96 - 1 -----------------------GCACUGCCAGCUAUCGAUAGCAAGUAAUCGGCUGGUCGUGAUGAAAUGGAUAAAAUUGCUUCCACUCUCUGGUAACACUGCAUUGUAUACCAGAAA- -----------------------.(((.((((((...((((........)))))))))).)))......((((..........))))...((((((((((......))).)))))))..- ( -28.20) >DroYak_CAF1 13818 109 - 1 ----------UCAGUAUUUUCCGGCACCGCCAGCUAUCGAUAGAAAAUAAUCGGCUGGUCGUGAUGAAUAAGA-AAAAAUGUUUUCACAACCUGGUAACACUGCAUUGUAUACCAGAAAA ----------....(((((.....(((.((((((...((((........)))))))))).)))..))))).((-(((....))))).....(((((((((......))).)))))).... ( -27.00) >consensus ___________C__UAU_U_C__GCACUGCCAGCUAUCGAUAGCAAAUAAUCGGCUGGUCGUGAUGAAAUGGA_AAAAUUGCUUCCACACUCUGGUAACACUGCAUUGUAUACCAGAAA_ ........................(((.((((((...((((........)))))))))).)))...........................((((((((((......))).)))))))... (-24.07 = -24.40 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:09 2006