
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,776,427 – 16,776,546 |
| Length | 119 |
| Max. P | 0.912154 |

| Location | 16,776,427 – 16,776,546 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.24 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
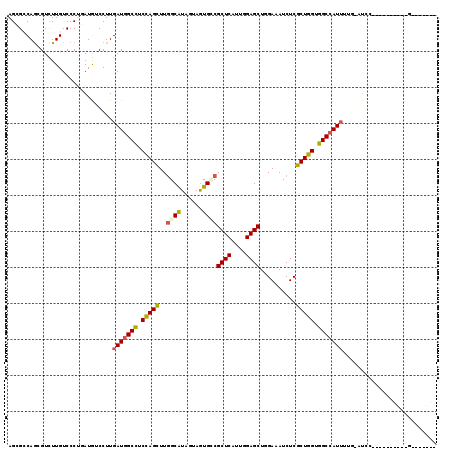
>X_DroMel_CAF1 16776427 119 + 22224390 AGCGACAGCGUCUUGUCCCUGAUGUCCUUGAUGGCCUCCAGUUUGGCGUAGUAGUGCCGCUCAUUGGAGCUCGAGAUCUCGCUGGUGGCCAUUGUAGAUCCAAAACGAUCCGUGUGGUA .((.(((.((..((((...((..(((..((((((((.(((((..((((......))))((((....))))..........))))).))))))))..))).))..))))..))))).)). ( -41.60) >DroPse_CAF1 55230 109 + 1 AGGGCCAGCGUCUUGUCCCGUAUGUCCUUGAUGGCCUCUAGCUUGGCAUAGUAGUGUCGCUCGUUGGAGCUGGAUAUCUCGCUGGUGGCCAUUUUGUGUCC----------GUGCGGGC (((((....)))))..((((((((..(..(((((((.(((((..(((((....)))))((((....))))..........))))).)))))))....)..)----------))))))). ( -46.50) >DroGri_CAF1 58446 102 + 1 AGCGCCAGUGUCUUGUCCCUGAUAUCCUUGAUUGCCUCCAGCUUGGCAUAAUAAUGCCGCUCGUUGGAGCUGGAAAUCUCGCUGGUGGCCAUUUUGUAUGC----------C------- .(((...(((((........))))).(..(((.(((.(((((..(((((....)))))((((....))))..........))))).))).)))..)..)))----------.------- ( -32.60) >DroEre_CAF1 15920 92 + 1 AGCGACAGGGUCUUGUCCCUGAUGUCCUUGAUGGCCUCCAGUUUGGCAUAGUAGUGACGCUCAUUGGAGCUCGAUAUCUCGCUGGUGGCCAU--------------------------- .(((.(((((......))))).))).....((((((.(((((..((.(((....(((.((((....))))))).))))).))))).))))))--------------------------- ( -35.70) >DroMoj_CAF1 77967 102 + 1 AGGGCCAACGUUUUGUCCCGUAUAUCCUUGAUGGCCUCCAGCUUAGCGUAGUAGUGCCGCUCAUUGGAGCUAGAAAUCUCGCUGGUGGCCAUUUUGAAUGG----------C------- .(((.(((....))).)))...(((.(..(((((((.(((((...(((......))).((((....))))..........))))).)))))))..).))).----------.------- ( -35.60) >DroAna_CAF1 58742 115 + 1 AGGGCCAGGGUCUUGUCCCUAAUGUCCUUGAUGGCUUCCAGCUUGGCGUAGUAGUGGCGCUCGUUGGAGCUGGAAAUCUCGCUGGUGGCCAUUU----UCUAAAUCGCCCUGUGUGGGA (((((.((((......))))...))))).(((((((.(((((..(((((.......))))).....(((.(....).)))))))).))))))).----.........(((.....))). ( -41.80) >consensus AGCGCCAGCGUCUUGUCCCUGAUGUCCUUGAUGGCCUCCAGCUUGGCAUAGUAGUGCCGCUCAUUGGAGCUGGAAAUCUCGCUGGUGGCCAUUUUG_AUCC__________G_______ .............................(((((((.(((((..((((......))))((((....))))..........))))).))))))).......................... (-26.58 = -26.58 + 0.00)

| Location | 16,776,427 – 16,776,546 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.24 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -19.40 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
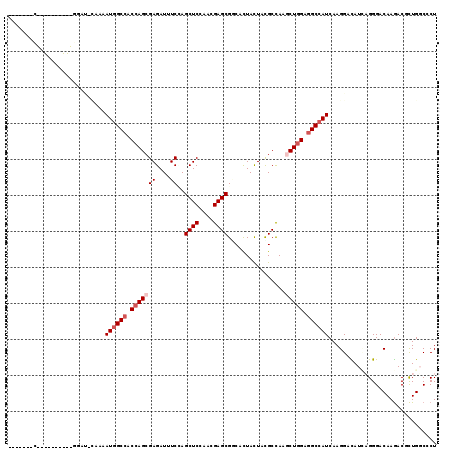
>X_DroMel_CAF1 16776427 119 - 22224390 UACCACACGGAUCGUUUUGGAUCUACAAUGGCCACCAGCGAGAUCUCGAGCUCCAAUGAGCGGCACUACUACGCCAAACUGGAGGCCAUCAAGGACAUCAGGGACAAGACGCUGUCGCU ..((....(((((......)))))...((((((.(((((((....))).((((....))))(((........)))...)))).))))))...)).....((.((((......)))).)) ( -39.70) >DroPse_CAF1 55230 109 - 1 GCCCGCAC----------GGACACAAAAUGGCCACCAGCGAGAUAUCCAGCUCCAACGAGCGACACUACUAUGCCAAGCUAGAGGCCAUCAAGGACAUACGGGACAAGACGCUGGCCCU (((.((..----------.........((((((.(.(((((....))..((((....))))................))).).))))))...(..(.....)..).....)).)))... ( -26.70) >DroGri_CAF1 58446 102 - 1 -------G----------GCAUACAAAAUGGCCACCAGCGAGAUUUCCAGCUCCAACGAGCGGCAUUAUUAUGCCAAGCUGGAGGCAAUCAAGGAUAUCAGGGACAAGACACUGGCGCU -------(----------((..........(((.(((((..........((((....))))(((((....)))))..))))).)))...........(((((.......).)))).))) ( -32.80) >DroEre_CAF1 15920 92 - 1 ---------------------------AUGGCCACCAGCGAGAUAUCGAGCUCCAAUGAGCGUCACUACUAUGCCAAACUGGAGGCCAUCAAGGACAUCAGGGACAAGACCCUGUCGCU ---------------------------((((((.(((((((....))).((((....)))).................)))).))))))...((.(..(((((......)))))..))) ( -30.80) >DroMoj_CAF1 77967 102 - 1 -------G----------CCAUUCAAAAUGGCCACCAGCGAGAUUUCUAGCUCCAAUGAGCGGCACUACUACGCUAAGCUGGAGGCCAUCAAGGAUAUACGGGACAAAACGUUGGCCCU -------(----------((((((...((((((.(((((..........((((....))))(((........)))..))))).))))))...))))..(((........))).)))... ( -35.40) >DroAna_CAF1 58742 115 - 1 UCCCACACAGGGCGAUUUAGA----AAAUGGCCACCAGCGAGAUUUCCAGCUCCAACGAGCGCCACUACUACGCCAAGCUGGAAGCCAUCAAGGACAUUAGGGACAAGACCCUGGCCCU ........(((((........----..(((((..(((((..........((((....))))((.........))...)))))..))))).........(((((......)))))))))) ( -35.70) >consensus _______C__________GGAU_CAAAAUGGCCACCAGCGAGAUUUCCAGCUCCAACGAGCGGCACUACUACGCCAAGCUGGAGGCCAUCAAGGACAUCAGGGACAAGACGCUGGCCCU ...........................((((((.(((((((....))..((((....))))................))))).)))))).............................. (-19.40 = -20.23 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:07 2006