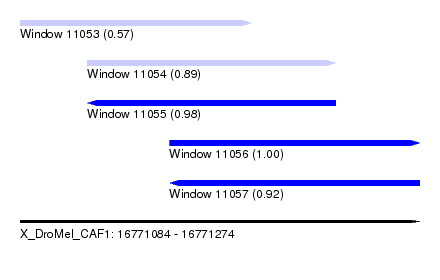
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,771,084 – 16,771,274 |
| Length | 190 |
| Max. P | 0.998437 |

| Location | 16,771,084 – 16,771,194 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16771084 110 + 22224390 CCUUUGUGUGUUAGUCUAGGUGUAAGCACACUCGUACUCGGUUGAAACGGACUUCCUGUUUUCAAAUAUAAAUCAAUCGAGACCUAUCGAAAAA-AUCGAUAUAAUCGAUC .....(((((((...(.....)..)))))))(((..((((((((((((((.....)))))............)))))))))...((((((....-.))))))....))).. ( -25.80) >DroSec_CAF1 8097 110 + 1 CCUUUGUGUGUUAGUCUAGGUGUAAGCACACUCGUACUCGGUUGAAACGGACUUCCUGUUUUCAAAUAUAAAUCAAUCGAGACCUAUCGAAAAA-UUCGAUAUAAUCGAUC .....(((((((...(.....)..)))))))(((..((((((((((((((.....)))))............)))))))))...((((((....-.))))))....))).. ( -25.80) >DroSim_CAF1 8027 110 + 1 CCUUUGUGUGUUAGUCUAGGUGUAAGCACACUCGUACUCGGUUGAAACGGACUUCCUGUUUUCAAAUAUAAAUCAAUCGAGACCUAUCGAAAAA-AUCGAUAUAAUCGAUC .....(((((((...(.....)..)))))))(((..((((((((((((((.....)))))............)))))))))...((((((....-.))))))....))).. ( -25.80) >DroEre_CAF1 10778 110 + 1 CCUUUGUGUGUUAGUCUAGGUGUAAGCACACUCGUGCCUAGUUGAUACGGACUUCCUGUUUUCAAAUAUAAAUUAAUCGACACCUAUCGAAAAC-AUCGAUAUAAUCGAUC ..((((.(((((((.(((((((..((....))..))))))))))))))((....))......)))).........(((((....((((((....-.))))))...))))). ( -26.50) >DroYak_CAF1 8018 111 + 1 CCUUUGUGUGUUAGUCUAGGUGUAAGCACACUUGUACUUGGUUGAUACGGACUUCCUGCUUUCAAAUAUAAAUUAAUCGACGCUUAUCGAAAUCGAUCGAUAUAAUCCAUC .......(((((((.(((((((((((....))))))))))))))))))(((......((......................)).((((((......))))))...)))... ( -21.95) >consensus CCUUUGUGUGUUAGUCUAGGUGUAAGCACACUCGUACUCGGUUGAAACGGACUUCCUGUUUUCAAAUAUAAAUCAAUCGAGACCUAUCGAAAAA_AUCGAUAUAAUCGAUC .....(((((((...(.....)..)))))))..((.(((((((((...((....))((....))........))))))))))).((((((......))))))......... (-19.80 = -19.96 + 0.16)

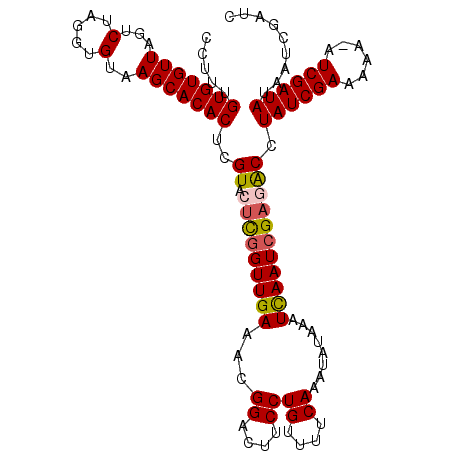
| Location | 16,771,116 – 16,771,234 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.38 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16771116 118 + 22224390 CGUACUCGGUUGAAACGGACUUCCUGUUUUCAAAUAUAAAUCAAUCGAGACCUAUCGAAAAA-AUCGAUAUAAUCGAUCGACUUAAAUUCGCAAGUGUUUUUUAUUUGCUGAAUGCAAG .((.((((((((((((((.....)))))............)))))))))))...((((....-.(((((...)))))))))(((..(((((((((((.....))))))).))))..))) ( -27.80) >DroSec_CAF1 8129 118 + 1 CGUACUCGGUUGAAACGGACUUCCUGUUUUCAAAUAUAAAUCAAUCGAGACCUAUCGAAAAA-UUCGAUAUAAUCGAUCGACUUAAAAUCGCAAGUGUUUUUUAUUUGCUGAAUGAAAG .((.((((((((((((((.....)))))............))))))))))).((((((....-.))))))...(((.((((.......))(((((((.....))))))).)).)))... ( -25.20) >DroSim_CAF1 8059 118 + 1 CGUACUCGGUUGAAACGGACUUCCUGUUUUCAAAUAUAAAUCAAUCGAGACCUAUCGAAAAA-AUCGAUAUAAUCGAUCGACUUAAAAUCGCAAGUGUUUUUUAUUUGCUGAAUGAAAG .((.((((((((((((((.....)))))............))))))))))).((((((....-.))))))...(((.((((.......))(((((((.....))))))).)).)))... ( -25.20) >DroEre_CAF1 10810 118 + 1 CGUGCCUAGUUGAUACGGACUUCCUGUUUUCAAAUAUAAAUUAAUCGACACCUAUCGAAAAC-AUCGAUAUAAUCGAUCGACUUGAAAUCGCAAAUGUUUUUUAUUUGCUGAAUGCAAG .........((((.((((.....))))..)))).......(..(((((....((((((....-.))))))...)))))..)((((...(((((((((.....))))))).))...)))) ( -24.00) >DroYak_CAF1 8050 119 + 1 UGUACUUGGUUGAUACGGACUUCCUGCUUUCAAAUAUAAAUUAAUCGACGCUUAUCGAAAUCGAUCGAUAUAAUCCAUCGACUUGAAAUCGCAAAUGUUUUUUAUUUGCCGAAUGCAAG ((((.(((((.((((.((((....(((((((((...........((((......))))..(((((.(((...))).))))).))))))..)))...))))..)))).))))).)))).. ( -24.80) >consensus CGUACUCGGUUGAAACGGACUUCCUGUUUUCAAAUAUAAAUCAAUCGAGACCUAUCGAAAAA_AUCGAUAUAAUCGAUCGACUUAAAAUCGCAAGUGUUUUUUAUUUGCUGAAUGCAAG .(((.(((.(((((((((.....))).))))))...(((.((.(((((....((((((......))))))...))))).)).))).....(((((((.....)))))))))).)))... (-21.62 = -21.38 + -0.24)

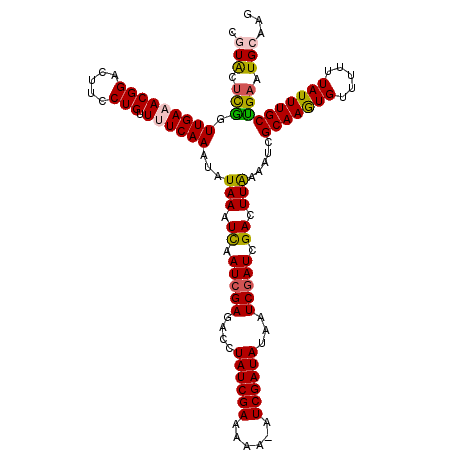
| Location | 16,771,116 – 16,771,234 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16771116 118 - 22224390 CUUGCAUUCAGCAAAUAAAAAACACUUGCGAAUUUAAGUCGAUCGAUUAUAUCGAU-UUUUUCGAUAGGUCUCGAUUGAUUUAUAUUUGAAAACAGGAAGUCCGUUUCAACCGAGUACG .((((.....)))).........(((((.((((.((((((((((((...((((((.-....))))))....)))))))))))).))))((((...((....)).))))...)))))... ( -32.80) >DroSec_CAF1 8129 118 - 1 CUUUCAUUCAGCAAAUAAAAAACACUUGCGAUUUUAAGUCGAUCGAUUAUAUCGAA-UUUUUCGAUAGGUCUCGAUUGAUUUAUAUUUGAAAACAGGAAGUCCGUUUCAACCGAGUACG .....((((.((((...........)))).....((((((((((((...((((((.-....))))))....))))))))))))...((((((...((....)).))))))..))))... ( -29.30) >DroSim_CAF1 8059 118 - 1 CUUUCAUUCAGCAAAUAAAAAACACUUGCGAUUUUAAGUCGAUCGAUUAUAUCGAU-UUUUUCGAUAGGUCUCGAUUGAUUUAUAUUUGAAAACAGGAAGUCCGUUUCAACCGAGUACG .....((((.((((...........)))).....((((((((((((...((((((.-....))))))....))))))))))))...((((((...((....)).))))))..))))... ( -30.10) >DroEre_CAF1 10810 118 - 1 CUUGCAUUCAGCAAAUAAAAAACAUUUGCGAUUUCAAGUCGAUCGAUUAUAUCGAU-GUUUUCGAUAGGUGUCGAUUAAUUUAUAUUUGAAAACAGGAAGUCCGUAUCAACUAGGCACG ..(((.....((((((.......))))))((((((((((..((((((..((((((.-....))))))...))))))..))))....(((....)))))))))............))).. ( -28.90) >DroYak_CAF1 8050 119 - 1 CUUGCAUUCGGCAAAUAAAAAACAUUUGCGAUUUCAAGUCGAUGGAUUAUAUCGAUCGAUUUCGAUAAGCGUCGAUUAAUUUAUAUUUGAAAGCAGGAAGUCCGUAUCAACCAAGUACA (((((.((((((((((.......)))))).......((((((((.....((((((......))))))..))))))))..........)))).)))))......((((.......)))). ( -27.90) >consensus CUUGCAUUCAGCAAAUAAAAAACACUUGCGAUUUUAAGUCGAUCGAUUAUAUCGAU_UUUUUCGAUAGGUCUCGAUUGAUUUAUAUUUGAAAACAGGAAGUCCGUUUCAACCGAGUACG .((((.....)))).........(((((.(.....(((((((((((...((((((......))))))....)))))))))))....((((((...((....)).))))))))))))... (-23.10 = -23.90 + 0.80)



| Location | 16,771,155 – 16,771,274 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.18 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16771155 119 + 22224390 AUCAAUCGAGACCUAUCGAAAAA-AUCGAUAUAAUCGAUCGACUUAAAUUCGCAAGUGUUUUUUAUUUGCUGAAUGCAAGUCAAACAAAUAGUUUCUGAAAUAUGCAUUUCGAAAUUCAA ....(((((....((((((....-.))))))...))))).(((((..(((((((((((.....))))))).))))..)))))........((((((.(((((....)))))))))))... ( -29.90) >DroSec_CAF1 8168 119 + 1 AUCAAUCGAGACCUAUCGAAAAA-UUCGAUAUAAUCGAUCGACUUAAAAUCGCAAGUGUUUUUUAUUUGCUGAAUGAAAGUCAAACAAACAGUUUCUGAAAUAUGCAUUUUGAUAUUCAA (((((((((....((((((....-.))))))...))))..(((((..(.(((((((((.....))))))).)).)..)))))...........................)))))...... ( -23.40) >DroSim_CAF1 8098 119 + 1 AUCAAUCGAGACCUAUCGAAAAA-AUCGAUAUAAUCGAUCGACUUAAAAUCGCAAGUGUUUUUUAUUUGCUGAAUGAAAGUCAAACAAACAGUUUCUGAAAUAUGCAUUUUGAUAUUCAA (((((((((....((((((....-.))))))...))))..(((((..(.(((((((((.....))))))).)).)..)))))...........................)))))...... ( -23.40) >DroEre_CAF1 10849 119 + 1 AUUAAUCGACACCUAUCGAAAAC-AUCGAUAUAAUCGAUCGACUUGAAAUCGCAAAUGUUUUUUAUUUGCUGAAUGCAAGUCAAACAUACAAUUUCUGAGAUAUGCAUUUCGAAAUUCAA ....(((((....((((((....-.))))))...))))).((((((...(((((((((.....))))))).))...))))))........((((((.(((((....)))))))))))... ( -29.20) >DroYak_CAF1 8089 115 + 1 AUUAAUCGACGCUUAUCGAAAUCGAUCGAUAUAAUCCAUCGACUUGAAAUCGCAAAUGUUUUUUAUUUGCCGAAUGCAAGUCAAACAUACAAUUUCUGAAAUAUGCAUUUCGAUA----- ....(((((.((.((((((......)))))).........((((((...(((((((((.....)))))).)))...))))))......................))...))))).----- ( -26.20) >consensus AUCAAUCGAGACCUAUCGAAAAA_AUCGAUAUAAUCGAUCGACUUAAAAUCGCAAGUGUUUUUUAUUUGCUGAAUGCAAGUCAAACAAACAGUUUCUGAAAUAUGCAUUUCGAUAUUCAA ....(((((....((((((......))))))...))))).(((((..(.(((((((((.....))))))).)).)..)))))...............(((((....)))))......... (-21.62 = -21.18 + -0.44)



| Location | 16,771,155 – 16,771,274 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16771155 119 - 22224390 UUGAAUUUCGAAAUGCAUAUUUCAGAAACUAUUUGUUUGACUUGCAUUCAGCAAAUAAAAAACACUUGCGAAUUUAAGUCGAUCGAUUAUAUCGAU-UUUUUCGAUAGGUCUCGAUUGAU ((((((((.(((.((((..((.((((.....))))...))..))))))).((((...........))))))))))))(((((((((...((((((.-....))))))....))))))))) ( -28.70) >DroSec_CAF1 8168 119 - 1 UUGAAUAUCAAAAUGCAUAUUUCAGAAACUGUUUGUUUGACUUUCAUUCAGCAAAUAAAAAACACUUGCGAUUUUAAGUCGAUCGAUUAUAUCGAA-UUUUUCGAUAGGUCUCGAUUGAU .((((..(((((..(((............)))...)))))..))))....((((...........))))........(((((((((...((((((.-....))))))....))))))))) ( -25.00) >DroSim_CAF1 8098 119 - 1 UUGAAUAUCAAAAUGCAUAUUUCAGAAACUGUUUGUUUGACUUUCAUUCAGCAAAUAAAAAACACUUGCGAUUUUAAGUCGAUCGAUUAUAUCGAU-UUUUUCGAUAGGUCUCGAUUGAU .((((..(((((..(((............)))...)))))..))))....((((...........))))........(((((((((...((((((.-....))))))....))))))))) ( -25.80) >DroEre_CAF1 10849 119 - 1 UUGAAUUUCGAAAUGCAUAUCUCAGAAAUUGUAUGUUUGACUUGCAUUCAGCAAAUAAAAAACAUUUGCGAUUUCAAGUCGAUCGAUUAUAUCGAU-GUUUUCGAUAGGUGUCGAUUAAU ...((((((...............))))))........((((((...((.((((((.......))))))))...))))))(((((((..((((((.-....))))))...)))))))... ( -29.96) >DroYak_CAF1 8089 115 - 1 -----UAUCGAAAUGCAUAUUUCAGAAAUUGUAUGUUUGACUUGCAUUCGGCAAAUAAAAAACAUUUGCGAUUUCAAGUCGAUGGAUUAUAUCGAUCGAUUUCGAUAAGCGUCGAUUAAU -----.(((((...((.((((...((((((((((((((.(.((((.....)))).)...))))))..))))))))((((((((.(((...))).)))))))).)))).)).))))).... ( -30.90) >consensus UUGAAUAUCGAAAUGCAUAUUUCAGAAACUGUUUGUUUGACUUGCAUUCAGCAAAUAAAAAACACUUGCGAUUUUAAGUCGAUCGAUUAUAUCGAU_UUUUUCGAUAGGUCUCGAUUGAU .........((((((((............((((((((.((.......)))))))))).........))).)))))..(((((((((...((((((......))))))....))))))))) (-20.64 = -21.04 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:06 2006