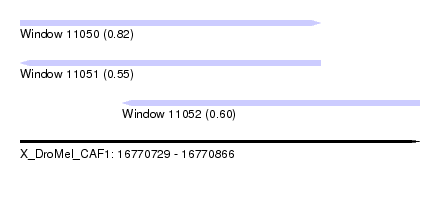
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,770,729 – 16,770,866 |
| Length | 137 |
| Max. P | 0.821926 |

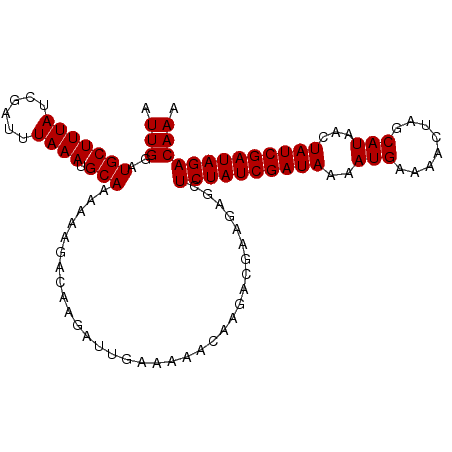
| Location | 16,770,729 – 16,770,832 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -18.17 |
| Consensus MFE | -15.83 |
| Energy contribution | -15.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16770729 103 + 22224390 AUUGGAUGCUUUAUCGAUUUAAAUGCAAAAAAGACAAGAUUGAAAAACAAGACGAAGAGCUCUAUCGAUAAAAUGAAAACUAGCAUAACUAUCGAUAGACAAA .(((...(((((.(((.......(((......).))...(((.....)))..))))))))((((((((((..(((........)))...))))))))))))). ( -17.60) >DroSec_CAF1 7742 103 + 1 AUUGGAUGCUUUAUCGAUUUAAAUGCAAAAAAGACAAGAUUGAAAAACAAGACGAAGAGGUCUAUCGAUAAAAUGAAAACUAGCAUAACUAUCGAUAGACAAA .(((..(..(((.(((((((...(((......).))))))))).)))..)..)))....(((((((((((..(((........)))...)))))))))))... ( -19.40) >DroSim_CAF1 7672 103 + 1 AUUGGAUGCUUUAUCGAUUUAAAUGCAAAAAAGACAAGAUUGAAAAACAAGACGAAAAGCUCUAUCGAUAAAAUGAUAACUAGCAUAACUAUCGAUAGACAAA .(((...(((((.(((.......(((......).))...(((.....)))..))))))))((((((((((..(((........)))...))))))))))))). ( -17.50) >consensus AUUGGAUGCUUUAUCGAUUUAAAUGCAAAAAAGACAAGAUUGAAAAACAAGACGAAGAGCUCUAUCGAUAAAAUGAAAACUAGCAUAACUAUCGAUAGACAAA .(((..(((((((......)))).))).................................((((((((((..(((........)))...))))))))))))). (-15.83 = -15.83 + 0.00)

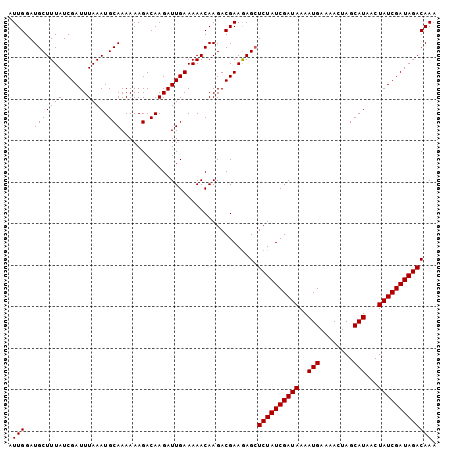
| Location | 16,770,729 – 16,770,832 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -18.34 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16770729 103 - 22224390 UUUGUCUAUCGAUAGUUAUGCUAGUUUUCAUUUUAUCGAUAGAGCUCUUCGUCUUGUUUUUCAAUCUUGUCUUUUUUGCAUUUAAAUCGAUAAAGCAUCCAAU ....(((((((((((..(((........))).)))))))))))(((..(((..(((.....(((...........)))....)))..)))...)))....... ( -17.70) >DroSec_CAF1 7742 103 - 1 UUUGUCUAUCGAUAGUUAUGCUAGUUUUCAUUUUAUCGAUAGACCUCUUCGUCUUGUUUUUCAAUCUUGUCUUUUUUGCAUUUAAAUCGAUAAAGCAUCCAAU ...((((((((((((..(((........))).))))))))))))......(..(((((.........(((.......)))........)))))..)....... ( -17.73) >DroSim_CAF1 7672 103 - 1 UUUGUCUAUCGAUAGUUAUGCUAGUUAUCAUUUUAUCGAUAGAGCUUUUCGUCUUGUUUUUCAAUCUUGUCUUUUUUGCAUUUAAAUCGAUAAAGCAUCCAAU ....(((((((((((..(((........))).)))))))))))((((((((..(((.....(((...........)))....)))..))).)))))....... ( -19.60) >consensus UUUGUCUAUCGAUAGUUAUGCUAGUUUUCAUUUUAUCGAUAGAGCUCUUCGUCUUGUUUUUCAAUCUUGUCUUUUUUGCAUUUAAAUCGAUAAAGCAUCCAAU ....(((((((((((..(((........))).)))))))))))(((..(((..(((.....(((...........)))....)))..)))...)))....... (-15.33 = -15.67 + 0.33)

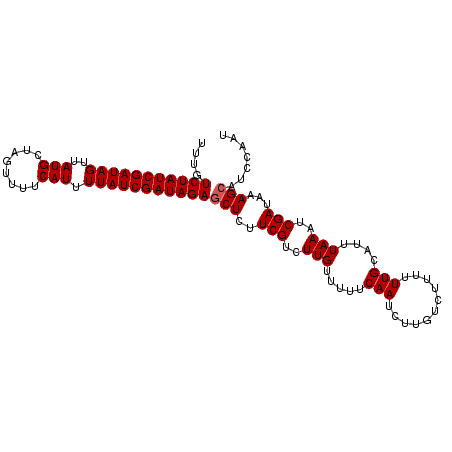
| Location | 16,770,764 – 16,770,866 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 90.08 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -17.14 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16770764 102 - 22224390 AUCGGCGCCGGUUGCUUAUCGAUCACGGUGCGUAUUUGUCUAUCGAUAGUUAUGCUAGUUUUCAUUUUAUCGAUAGAGCUCUUCGUCUUGUUU-UUCAAUCUU ....((((((((((.....))))..)))))).......(((((((((((..(((........))).)))))))))))................-......... ( -23.40) >DroSec_CAF1 7777 102 - 1 AUCGGCACUGGUUGCUUAUCGAUCGCGGUGCGUAUUUGUCUAUCGAUAGUUAUGCUAGUUUUCAUUUUAUCGAUAGACCUCUUCGUCUUGUUU-UUCAAUCUU ....((((((((((.....)))))..)))))......((((((((((((..(((........))).))))))))))))...............-......... ( -24.20) >DroSim_CAF1 7707 102 - 1 AUCGGCACUGGUUGCUUAUCGAUCGCGGUGCGUAUUUGUCUAUCGAUAGUUAUGCUAGUUAUCAUUUUAUCGAUAGAGCUUUUCGUCUUGUUU-UUCAAUCUU ....((((((((((.....)))))..))))).......(((((((((((..(((........))).)))))))))))................-......... ( -21.70) >DroEre_CAF1 10456 103 - 1 AUCGGCGCUAGUUGUUUAUCGAUCACGGAGCGUAUUUGUCUAUCGAUAGGCAUGUUCGUUUUCUCUAUAUCAAUAGAACUCUUCGUCUUUUUUCUUCAAUCUU ...((((..((((((((((((((.(((((.....)))))..)))))))))))...........(((((....))))))))...))))................ ( -20.60) >consensus AUCGGCACUGGUUGCUUAUCGAUCACGGUGCGUAUUUGUCUAUCGAUAGUUAUGCUAGUUUUCAUUUUAUCGAUAGAGCUCUUCGUCUUGUUU_UUCAAUCUU ....((((((((((.....))))..)))))).......((((((((((...(((........)))..)))))))))).......................... (-17.14 = -18.07 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:01 2006