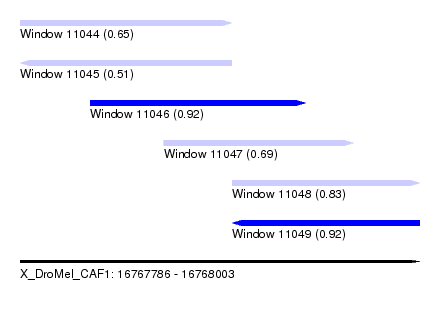
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,767,786 – 16,768,003 |
| Length | 217 |
| Max. P | 0.923384 |

| Location | 16,767,786 – 16,767,901 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 96.51 |
| Mean single sequence MFE | -12.29 |
| Consensus MFE | -11.82 |
| Energy contribution | -12.15 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16767786 115 + 22224390 AAAACUAAAAACCAAUAACUUAAAACUAAUUACAUUAUUAAGCAACUUUUUCCUCAGCUUCAUUAAUUUCCCAAAAUUUAUCGCACAUCAUUGCUCGAAUGAUGUGUUAUCGAUA .................................((((..((((.............))))...))))...........((((((((((((((.....)))))))))....))))) ( -13.72) >DroSec_CAF1 5206 114 + 1 UAAACUAAAUACCAAUAACUUAAAACUAAUUACAUUAUUAAGCAACUUUUUCCUCAGCUUCAUUAAUUUCCCAAA-UUUAUCGCACAUCAUUUCUCGAAUGAUGUGUUAUCGAUA .................................((((..((((.............))))...))))........-..(((((((((((((((...))))))))))....))))) ( -13.72) >DroSim_CAF1 5130 113 + 1 UAAACUAAAUACCAAUAACUUAAAACUAAUUACAUUAUUAAGCAACUUUUUCCUCAGCUUCAUUAAUUUCCCAAA-U-UAUCGCCCAUCAUUGCUCGAAUGAUGUGUUAUCGAUA ............(.(((((..............((((..((((.............))))...))))........-.-.......(((((((.....))))))).))))).)... ( -9.42) >consensus UAAACUAAAUACCAAUAACUUAAAACUAAUUACAUUAUUAAGCAACUUUUUCCUCAGCUUCAUUAAUUUCCCAAA_UUUAUCGCACAUCAUUGCUCGAAUGAUGUGUUAUCGAUA .................................((((..((((.............))))...))))...........((((((((((((((.....)))))))))....))))) (-11.82 = -12.15 + 0.33)



| Location | 16,767,786 – 16,767,901 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 96.51 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16767786 115 - 22224390 UAUCGAUAACACAUCAUUCGAGCAAUGAUGUGCGAUAAAUUUUGGGAAAUUAAUGAAGCUGAGGAAAAAGUUGCUUAAUAAUGUAAUUAGUUUUAAGUUAUUGGUUUUUAGUUUU (((((....(((((((((.....)))))))))))))).................(((((((((((...(((.((((((...((....))...)))))).)))..))))))))))) ( -25.60) >DroSec_CAF1 5206 114 - 1 UAUCGAUAACACAUCAUUCGAGAAAUGAUGUGCGAUAAA-UUUGGGAAAUUAAUGAAGCUGAGGAAAAAGUUGCUUAAUAAUGUAAUUAGUUUUAAGUUAUUGGUAUUUAGUUUA ((((((((((((((((((.....))))))))........-.............((((((((........(((((........))))))))))))).))))))))))......... ( -23.30) >DroSim_CAF1 5130 113 - 1 UAUCGAUAACACAUCAUUCGAGCAAUGAUGGGCGAUA-A-UUUGGGAAAUUAAUGAAGCUGAGGAAAAAGUUGCUUAAUAAUGUAAUUAGUUUUAAGUUAUUGGUAUUUAGUUUA ((((((((((.(((((((.....))))))).......-.-.............((((((((........(((((........))))))))))))).))))))))))......... ( -22.30) >consensus UAUCGAUAACACAUCAUUCGAGCAAUGAUGUGCGAUAAA_UUUGGGAAAUUAAUGAAGCUGAGGAAAAAGUUGCUUAAUAAUGUAAUUAGUUUUAAGUUAUUGGUAUUUAGUUUA ((((((((((.(((((((.....))))))).......................((((((((........(((((........))))))))))))).))))))))))......... (-20.90 = -21.23 + 0.33)


| Location | 16,767,824 – 16,767,941 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -14.45 |
| Energy contribution | -15.95 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16767824 117 + 22224390 UAAGCAACUUUUUCCUCAGCUUCAUUAAUUUCCCAAAAUUUAUCGCACAUCAUU-GCUCGAAUGAUGUGUUAUCGAUAAAUUCGAAAUACGAAAUUCGAAAAACGGCUUGCAGAGCAA (((((...((((((.............(((((....((((((((((((((((((-.....)))))))))....))))))))).)))))..(.....)))))))..)))))........ ( -24.80) >DroSec_CAF1 5244 115 + 1 UAAGCAACUUUUUCCUCAGCUUCAUUAAUUUCCCAAA-UUUAUCGCACAUCAUU-UCUCGAAUGAUGUGUUAUCGAUAAAUUCGAUAUUCGAAAUUCAAAAAGC-GCUUGCAGAGCAA ..................((((....((((((.....-......((((((((((-(...)))))))))))((((((.....))))))...))))))....))))-((((...)))).. ( -26.80) >DroSim_CAF1 5168 114 + 1 UAAGCAACUUUUUCCUCAGCUUCAUUAAUUUCCCAAA-U-UAUCGCCCAUCAUU-GCUCGAAUGAUGUGUUAUCGAUAAAUUCGAUAUUCGAAAUUCAAAAAGC-GCUUGCAGAGCAA ..................((((....((((((.....-.-....((.(((((((-.....))))))).))((((((.....))))))...))))))....))))-((((...)))).. ( -22.00) >DroEre_CAF1 8160 99 + 1 UAAGCAACUUUUUCCUCAGCUUUAUUAAUUUCCCAA-AUUUAUCGCACAUCAUUCACUCGAAUGCU--GUUCUCGAUAAAUUCGAAAUGCA----------------CUCCAGAGCAA ..................(((((....(((((...(-((((((((.(((.(((((....))))).)--))...))))))))).)))))...----------------....))))).. ( -20.00) >consensus UAAGCAACUUUUUCCUCAGCUUCAUUAAUUUCCCAAA_UUUAUCGCACAUCAUU_GCUCGAAUGAUGUGUUAUCGAUAAAUUCGAAAUUCGAAAUUCAAAAAGC_GCUUGCAGAGCAA ..................((((......................((((((((((......))))))))))((((((.....)))))).........................)))).. (-14.45 = -15.95 + 1.50)


| Location | 16,767,864 – 16,767,967 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 84.95 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -13.05 |
| Energy contribution | -14.55 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16767864 103 + 22224390 UAUCGCACAUCAUU-GCUCGAAUGAUGUGUUAUCGAUAAAUUCGAAAUACGAAAUUCGAAAAACGGCUUGCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCC ((((((((((((((-.....)))))))))....)))))..((((((........))))))....(((((((...)))..))))..(((.........))).... ( -24.50) >DroSec_CAF1 5283 102 + 1 UAUCGCACAUCAUU-UCUCGAAUGAUGUGUUAUCGAUAAAUUCGAUAUUCGAAAUUCAAAAAGC-GCUUGCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCC ..((((((((((((-(...)))))))))))((((((.....))))))...)).........(((-((((...))))..((....)).))).............. ( -22.40) >DroSim_CAF1 5206 102 + 1 UAUCGCCCAUCAUU-GCUCGAAUGAUGUGUUAUCGAUAAAUUCGAUAUUCGAAAUUCAAAAAGC-GCUUGCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCC ....((.(((..((-((((((((..((...((((((.....))))))..))..)))).....((-....)).)))))).....))).))............... ( -21.30) >DroEre_CAF1 8199 86 + 1 UAUCGCACAUCAUUCACUCGAAUGCU--GUUCUCGAUAAAUUCGAAAUGCA----------------CUCCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCC ....((.(((.((((....))))(((--((..((((.....))))..(((.----------------.......)))))))).))).))............... ( -14.80) >consensus UAUCGCACAUCAUU_GCUCGAAUGAUGUGUUAUCGAUAAAUUCGAAAUUCGAAAUUCAAAAAGC_GCUUGCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCC ....((((((((((......))))))))))((((((.....))))))........................((((....(((.....))).......))))... (-13.05 = -14.55 + 1.50)


| Location | 16,767,901 – 16,768,003 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16767901 102 + 22224390 AAUUCGAAAUACGAAAUUCGAAAAACGGCUUGCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCCAUUACAGGGCACGGCCAAUCUGCAAUCCUCUAUAAA ..((((((........))))))....((.(((((((......(((..(((.........))).(((......)))...)))...))))))).))........ ( -25.20) >DroSec_CAF1 5320 101 + 1 AAUUCGAUAUUCGAAAUUCAAAAAGC-GCUUGCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCCAUUCCAGGGCACGGCCAAUCUGCAAUCCUCUAUAAA ..((((.....))))........((.-(.(((((((......(((..(((.........))).(((......)))...)))...))))))).)))....... ( -20.40) >DroSim_CAF1 5243 101 + 1 AAUUCGAUAUUCGAAAUUCAAAAAGC-GCUUGCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCCAUUCCAGGGCACGGCCAAUCUGCAAUCCUCUAUAAA ..((((.....))))........((.-(.(((((((......(((..(((.........))).(((......)))...)))...))))))).)))....... ( -20.40) >DroEre_CAF1 8235 86 + 1 AAUUCGAAAUGCA----------------CUCCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCCAUUCUAGGGCACGGCCAAUCUGCAAUCCUCUAUAAA .............----------------...((((......(((..(((.........))).(((......)))...)))...)))).............. ( -11.50) >consensus AAUUCGAAAUUCGAAAUUCAAAAAGC_GCUUGCAGAGCAACAGCCAUGAGCUAACCAAACUCUCCCAUUCCAGGGCACGGCCAAUCUGCAAUCCUCUAUAAA ..............................((((((......(((..(((.........))).(((......)))...)))...))))))............ (-15.15 = -15.40 + 0.25)


| Location | 16,767,901 – 16,768,003 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -24.65 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16767901 102 - 22224390 UUUAUAGAGGAUUGCAGAUUGGCCGUGCCCUGUAAUGGGAGAGUUUGGUUAGCUCAUGGCUGUUGCUCUGCAAGCCGUUUUUCGAAUUUCGUAUUUCGAAUU ........((.(((((((.(((((((.(((......))).(((((.....))))))))))))....))))))).))....((((((........)))))).. ( -34.30) >DroSec_CAF1 5320 101 - 1 UUUAUAGAGGAUUGCAGAUUGGCCGUGCCCUGGAAUGGGAGAGUUUGGUUAGCUCAUGGCUGUUGCUCUGCAAGC-GCUUUUUGAAUUUCGAAUAUCGAAUU ((((.(((((.(((((((.(((((((.(((......))).(((((.....))))))))))))....))))))).)-.)))).)))).((((.....)))).. ( -32.10) >DroSim_CAF1 5243 101 - 1 UUUAUAGAGGAUUGCAGAUUGGCCGUGCCCUGGAAUGGGAGAGUUUGGUUAGCUCAUGGCUGUUGCUCUGCAAGC-GCUUUUUGAAUUUCGAAUAUCGAAUU ((((.(((((.(((((((.(((((((.(((......))).(((((.....))))))))))))....))))))).)-.)))).)))).((((.....)))).. ( -32.10) >DroEre_CAF1 8235 86 - 1 UUUAUAGAGGAUUGCAGAUUGGCCGUGCCCUAGAAUGGGAGAGUUUGGUUAGCUCAUGGCUGUUGCUCUGGAG----------------UGCAUUUCGAAUU ........(.(((.((((.(((((((.(((......))).(((((.....))))))))))))....)))).))----------------).).......... ( -24.90) >consensus UUUAUAGAGGAUUGCAGAUUGGCCGUGCCCUGGAAUGGGAGAGUUUGGUUAGCUCAUGGCUGUUGCUCUGCAAGC_GCUUUUUGAAUUUCGAAUAUCGAAUU ...........(((((((.(((((((.(((......))).(((((.....))))))))))))....)))))))..............((((.....)))).. (-24.65 = -25.03 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:59 2006