
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,766,556 – 16,766,652 |
| Length | 96 |
| Max. P | 0.929031 |

| Location | 16,766,556 – 16,766,652 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.28 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -17.35 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.929031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
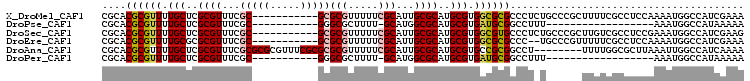
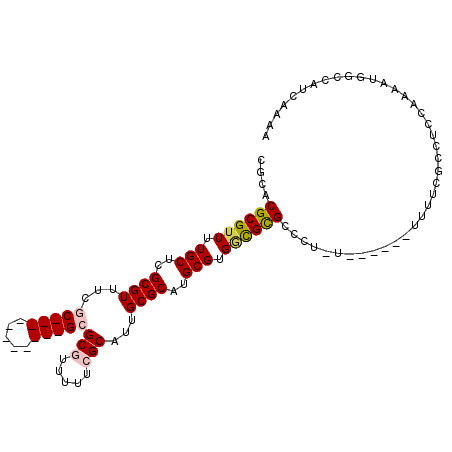
>X_DroMel_CAF1 16766556 96 + 22224390 UUUCGAUGGCCAUUUUGGAGGCGAAAAGCGGGCAGAGGGCGCGCCACGCAUGCGCAAUGCGAAAAACGCGC-----------GCGAAACGCGAGCAAAACGCGUGCG (((((....((.....))...))))).(((.((.....)).)))..(((((.....))))).....(((((-----------(((...(....).....)))))))) ( -34.20) >DroPse_CAF1 47954 77 + 1 UUUUUAUGGCCAUUU------------------AAAGGCCGCAUCACGCAUGCGCCAUGC-AAAAGCGCCC-----------GCGAAACGCGAGCAAAACGCGUGCG .......((((....------------------...))))......((((((((....((-....))((.(-----------(((...)))).))....)))))))) ( -29.20) >DroSec_CAF1 4038 96 + 1 CUUCGAUGGCCAUUUCGGAGGCGACAAGCGGGCAGAGGGCACGCCACGCAUGCGCAAUGCGAAAAACGCGC-----------GCGAAACGCGAGCAAAACGCGUGCG ((((((........))))))(((.((.((((((.........))).))).)))))...........(((((-----------(((...(....).....)))))))) ( -35.90) >DroEre_CAF1 6946 94 + 1 UUUCGAUGGCCAUUUUGGAGGCGAAAAACGGGCA--GGGCGCGCCACGCAUGCGCAAUGCGAAAAACGCGC-----------GCGAAACGCGCGCAAAACGCGUGCG (((((....((.....))...)))))........--..((((((..(((((.....)))))......((((-----------(((...))))))).....)))))). ( -37.30) >DroAna_CAF1 48812 99 + 1 UUUUGAUGGCCAAUUUAAGCGCCAAAA--------AGGCCGCGGCACGCAUGCGCAAUGCGAAAAACGCGCGCGAAACGCGCGCGAAACGCGAGCAAAACGCGUGCG ........(((.......(((((....--------.)).)))))).((((((((...(((......((((((((...))))))))........)))...)))))))) ( -39.55) >DroPer_CAF1 48518 77 + 1 UUUUUAUGGCCAUUU------------------AAAGGCCGCAUCACGCAUGCGCCAUGC-AAAAGCGCCC-----------GCGAAACGCGAGCAAAACGCGUGCG .......((((....------------------...))))......((((((((....((-....))((.(-----------(((...)))).))....)))))))) ( -29.20) >consensus UUUCGAUGGCCAUUUUGGAGGCGAAAA______A_AGGCCGCGCCACGCAUGCGCAAUGCGAAAAACGCGC___________GCGAAACGCGAGCAAAACGCGUGCG ......((.((.........................)).)).....((((((((...(((........((((((.....))))))........)))...)))))))) (-17.35 = -18.17 + 0.82)



| Location | 16,766,556 – 16,766,652 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.28 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -17.89 |
| Energy contribution | -18.82 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16766556 96 - 22224390 CGCACGCGUUUUGCUCGCGUUUCGC-----------GCGCGUUUUUCGCAUUGCGCAUGCGUGGCGCGCCCUCUGCCCGCUUUUCGCCUCCAAAAUGGCCAUCGAAA .(((.((((..(((.(((((.....-----------)))))......)))..)))).)))((((.(((.....))))))).....(((........)))........ ( -32.40) >DroPse_CAF1 47954 77 - 1 CGCACGCGUUUUGCUCGCGUUUCGC-----------GGGCGCUUUU-GCAUGGCGCAUGCGUGAUGCGGCCUUU------------------AAAUGGCCAUAAAAA ((((.(((((..(((((((...)))-----------))))((....-))..))))).))))......((((...------------------....))))....... ( -32.50) >DroSec_CAF1 4038 96 - 1 CGCACGCGUUUUGCUCGCGUUUCGC-----------GCGCGUUUUUCGCAUUGCGCAUGCGUGGCGUGCCCUCUGCCCGCUUGUCGCCUCCGAAAUGGCCAUCGAAG ((((.((((..(((.(((((.....-----------)))))......)))..)))).)))).((((.((.....)).))))....(((........)))........ ( -31.60) >DroEre_CAF1 6946 94 - 1 CGCACGCGUUUUGCGCGCGUUUCGC-----------GCGCGUUUUUCGCAUUGCGCAUGCGUGGCGCGCCC--UGCCCGUUUUUCGCCUCCAAAAUGGCCAUCGAAA ((((.(((....(((((((.....)-----------))))))....)))..))))...(((.((((.....--))))))).....(((........)))........ ( -33.70) >DroAna_CAF1 48812 99 - 1 CGCACGCGUUUUGCUCGCGUUUCGCGCGCGUUUCGCGCGCGUUUUUCGCAUUGCGCAUGCGUGCCGCGGCCU--------UUUUGGCGCUUAAAUUGGCCAUCAAAA .((((((((..(((..(((...((((((((...)))))))).....)))...))).))))))))...((((.--------.((((.....))))..))))....... ( -44.00) >DroPer_CAF1 48518 77 - 1 CGCACGCGUUUUGCUCGCGUUUCGC-----------GGGCGCUUUU-GCAUGGCGCAUGCGUGAUGCGGCCUUU------------------AAAUGGCCAUAAAAA ((((.(((((..(((((((...)))-----------))))((....-))..))))).))))......((((...------------------....))))....... ( -32.50) >consensus CGCACGCGUUUUGCUCGCGUUUCGC___________GCGCGUUUUUCGCAUUGCGCAUGCGUGGCGCGCCCU_U______UUUUCGCCUCCAAAAUGGCCAUCAAAA ....((((((.(((..((((...(((((.....)))))(((.....)))...))))..))).))))))....................................... (-17.89 = -18.82 + 0.93)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:54 2006