
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,765,540 – 16,765,648 |
| Length | 108 |
| Max. P | 0.575083 |

| Location | 16,765,540 – 16,765,648 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.01 |
| Mean single sequence MFE | -22.89 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
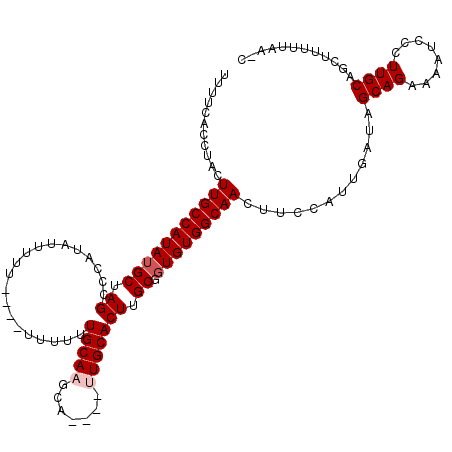
>X_DroMel_CAF1 16765540 108 + 22224390 UUUUCACCUACUUGCCAUAUGCUAGCCCAUAUUUUU----UUUUUUGCAUGCA----UUGCACUUGCGGUGUGGCAACUUCCAUUGAUAGCAGAAAAUCCCUUGCAGCUUUUUAA-C ...........(((((..((((..............----......))))(((----((((....))))))))))))............((((........))))..........-. ( -19.45) >DroSec_CAF1 3049 113 + 1 UUUUCACCUACUUGCCAUAUGCUAGCCCAUAUUUUU----UUUUUUGCAAGCAUUGAUUGCACUUGCGGUGUGGCAACUUGCAUUGAUAGCAGAAAAUCCCUUGCAGCUUUUUAACC ...........((((((((((((.((..........----......)).)))))..(((((....)))))))))))).(((((..(((........)))...))))).......... ( -21.59) >DroSim_CAF1 2955 117 + 1 UUUUCACCUACUUGCCAUAUGCUAGCCCAUAUUUUUUUUUUUUUUUGCAAGCAUUGAUUGCACUUGCGGUGUGGCAACUUGCAUUGAUAGCAGAAAAUCCCUUGCAGCUUUUUAACC ....................(((.((....(((((((.((.((..((((((..(((.(..(((.....)))..))))))))))..)).)).))))))).....)))))......... ( -21.80) >DroEre_CAF1 6144 104 + 1 UUCUCACCUACUUGCCAUAAGCUAGUCCAUAUUUAU--------UUGCACACA----UUGCACUUGCGGUGUGGCAACUGCAAGCGAUAGCAGUAAAUCCCUUGCAGCCUAUUAA-C .........(((.((.....)).)))..........--------((((.((((----((((....))))))))))))(((((((.(((........))).)))))))........-. ( -28.70) >consensus UUUUCACCUACUUGCCAUAUGCUAGCCCAUAUUUUU____UUUUUUGCAAGCA____UUGCACUUGCGGUGUGGCAACUUCCAUUGAUAGCAGAAAAUCCCUUGCAGCUUUUUAA_C ...........(((((((((((.((....................(((((.......))))))).)).)))))))))............((((........))))............ (-15.65 = -16.40 + 0.75)

| Location | 16,765,540 – 16,765,648 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.01 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.77 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16765540 108 - 22224390 G-UUAAAAAGCUGCAAGGGAUUUUCUGCUAUCAAUGGAAGUUGCCACACCGCAAGUGCAA----UGCAUGCAAAAAA----AAAAAUAUGGGCUAGCAUAUGGCAAGUAGGUGAAAA .-..............(((((((((..........))))))).)).(((((((.((....----.)).)))......----.....(((..((((.....))))..))))))).... ( -21.20) >DroSec_CAF1 3049 113 - 1 GGUUAAAAAGCUGCAAGGGAUUUUCUGCUAUCAAUGCAAGUUGCCACACCGCAAGUGCAAUCAAUGCUUGCAAAAAA----AAAAAUAUGGGCUAGCAUAUGGCAAGUAGGUGAAAA (((.....(((((((...(((........)))..))).))))))).((((((((((.........))))))......----.....(((..((((.....))))..))))))).... ( -27.40) >DroSim_CAF1 2955 117 - 1 GGUUAAAAAGCUGCAAGGGAUUUUCUGCUAUCAAUGCAAGUUGCCACACCGCAAGUGCAAUCAAUGCUUGCAAAAAAAAAAAAAAAUAUGGGCUAGCAUAUGGCAAGUAGGUGAAAA (((.....(((((((...(((........)))..))).))))))).((((((((((.........))))))...............(((..((((.....))))..))))))).... ( -27.40) >DroEre_CAF1 6144 104 - 1 G-UUAAUAGGCUGCAAGGGAUUUACUGCUAUCGCUUGCAGUUGCCACACCGCAAGUGCAA----UGUGUGCAA--------AUAAAUAUGGACUAGCUUAUGGCAAGUAGGUGAGAA .-......(((((((((.(((........))).)))))))))((((((..((....))..----)))).))..--------...........((.((((((.....)))))).)).. ( -30.20) >consensus G_UUAAAAAGCUGCAAGGGAUUUUCUGCUAUCAAUGCAAGUUGCCACACCGCAAGUGCAA____UGCUUGCAAAAAA____AAAAAUAUGGGCUAGCAUAUGGCAAGUAGGUGAAAA ........(.((((....(((........)))........(((((.....(((...(((.....))).)))..............(((((......)))))))))))))).)..... (-17.90 = -17.77 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:52 2006