
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,761,027 – 16,761,167 |
| Length | 140 |
| Max. P | 0.836242 |

| Location | 16,761,027 – 16,761,128 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
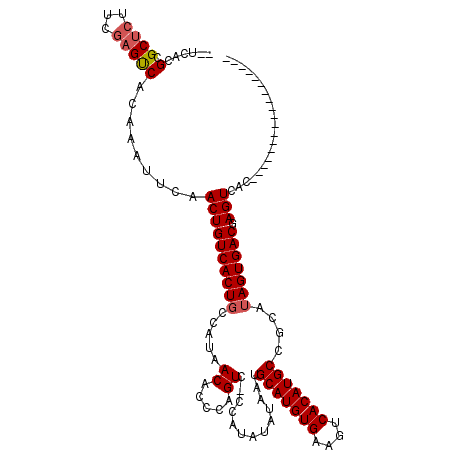
>X_DroMel_CAF1 16761027 101 + 22224390 GUUCACGCGCUCUUCGAGUCACAAAUUCAACUGUCACUGCCAUAACACCCAGUC-CCAUAUAUAAUGCAUGUGAAGUCACAUGCCAUAUAGUGACAAGUCAC------------------ .................(((((.........((..((((..........)))).-.))(((((...(((((((....))))))).)))))))))).......------------------ ( -21.60) >DroSec_CAF1 47822 99 + 1 --UCACGCGCUCUUCGAGUCACAAAUUCAACUGUCACUGCCAUAACACCCAGUC-CCAUAUAUAAUGCAUGUGAAGUCACAUGCCGCAAAGUGACGAGUCAC------------------ --......((((...((((.....))))....((((((((....((.....)).-...........(((((((....))))))).))..))))))))))...------------------ ( -22.40) >DroSim_CAF1 45751 99 + 1 --UCACGCGCUCUUCGAGUCACAAAUUCGACUGUCACUGCCAUAACACCCAGUC-CCAUAUAUAAUGCAUGUGAAGUCACAUGCCGCAUAGUGACGAGUCAC------------------ --......((((.((((((.....))))))..(((((((.....((.....)).-...........(((((((....)))))))....)))))))))))...------------------ ( -25.50) >DroYak_CAF1 40012 116 + 1 ----UCGUGCUCUUCACGCCGCAAAUUCAACUGUCACUGCCAUAACACCAAGUCCCCAUAUAUAAUGCAUGUGAAGUCACAUGCCGUAUAGUGACGAGUCACUCACUAGUCGAAUCGGAU ----.((((.....))))(((...((((.((((((((((.....((.....)).............(((((((....)))))))....)))))))(((...)))...))).))))))).. ( -27.30) >consensus __UCACGCGCUCUUCGAGUCACAAAUUCAACUGUCACUGCCAUAACACCCAGUC_CCAUAUAUAAUGCAUGUGAAGUCACAUGCCGCAUAGUGACGAGUCAC__________________ ......(.((((...))))).........((((((((((.....((.....)).............(((((((....)))))))....))))))).)))..................... (-17.94 = -18.50 + 0.56)

| Location | 16,761,027 – 16,761,128 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -28.41 |
| Energy contribution | -27.48 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16761027 101 - 22224390 ------------------GUGACUUGUCACUAUAUGGCAUGUGACUUCACAUGCAUUAUAUAUGG-GACUGGGUGUUAUGGCAGUGACAGUUGAAUUUGUGACUCGAAGAGCGCGUGAAC ------------------....((.(((.((((((((((((((....)))))))....)))))))-))).))(((((.....(((.((((......)))).))).....)))))...... ( -31.10) >DroSec_CAF1 47822 99 - 1 ------------------GUGACUCGUCACUUUGCGGCAUGUGACUUCACAUGCAUUAUAUAUGG-GACUGGGUGUUAUGGCAGUGACAGUUGAAUUUGUGACUCGAAGAGCGCGUGA-- ------------------(((.(((((((((..((.(((((((....)))))))......(((((-.((...)).)))))))))))))((((........))))....))))))....-- ( -30.10) >DroSim_CAF1 45751 99 - 1 ------------------GUGACUCGUCACUAUGCGGCAUGUGACUUCACAUGCAUUAUAUAUGG-GACUGGGUGUUAUGGCAGUGACAGUCGAAUUUGUGACUCGAAGAGCGCGUGA-- ------------------(((.(((((((((..((.(((((((....)))))))......(((((-.((...)).)))))))))))))(((((......)))))....))))))....-- ( -32.40) >DroYak_CAF1 40012 116 - 1 AUCCGAUUCGACUAGUGAGUGACUCGUCACUAUACGGCAUGUGACUUCACAUGCAUUAUAUAUGGGGACUUGGUGUUAUGGCAGUGACAGUUGAAUUUGCGGCGUGAAGAGCACGA---- ..(((((((((((.....((.(((.((((..((((.(((((((....))))))).........(....)...))))..))))))).))))))))))...)))((((.....)))).---- ( -36.60) >consensus __________________GUGACUCGUCACUAUACGGCAUGUGACUUCACAUGCAUUAUAUAUGG_GACUGGGUGUUAUGGCAGUGACAGUUGAAUUUGUGACUCGAAGAGCGCGUGA__ ..................(((.(((((((((.....(((((((....)))))))............(((.....))).....)))))).((((......)))).....))))))...... (-28.41 = -27.48 + -0.94)



| Location | 16,761,067 – 16,761,167 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.08 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16761067 100 + 22224390 CAUAACACCCAGUC-CCAUAUAUAAUGCAUGUGAAGUCACAUGCCAUAUAGUGACAAGUCAC-------------------GUAUUGCAUCACAAAAAAAUCUUAGACAACUGCCAGAGU ...........(((-...(((((...(((((((....))))))).)))))((((....))))-------------------........................)))............ ( -18.90) >DroSec_CAF1 47860 99 + 1 CAUAACACCCAGUC-CCAUAUAUAAUGCAUGUGAAGUCACAUGCCGCAAAGUGACGAGUCAC-------------------GGAUUGCAUCACAAA-ACAUCUUAGACAAUUGCCAGAGU ...........(((-...........(((((((....))))))).((((.((((....))))-------------------...))))........-........)))............ ( -21.90) >DroSim_CAF1 45789 99 + 1 CAUAACACCCAGUC-CCAUAUAUAAUGCAUGUGAAGUCACAUGCCGCAUAGUGACGAGUCAC-------------------GGAUUGCAUCACAAA-ACAUCUUAGACAAUUGCCAGAGU .........(((((-(..........(((((((....)))))))......((((....))))-------------------)))))).........-....................... ( -21.80) >DroYak_CAF1 40048 119 + 1 CAUAACACCAAGUCCCCAUAUAUAAUGCAUGUGAAGUCACAUGCCGUAUAGUGACGAGUCACUCACUAGUCGAAUCGGAUUGGAUUGCAUCAGAGA-AAAUCUUAGACAAUCGCCAGAGU ..........(((((...........(((((((....)))))))((..((((((........))))))..))....))))).(((((..((.(((.-....))).)))))))........ ( -27.90) >consensus CAUAACACCCAGUC_CCAUAUAUAAUGCAUGUGAAGUCACAUGCCGCAUAGUGACGAGUCAC___________________GGAUUGCAUCACAAA_AAAUCUUAGACAAUUGCCAGAGU ...........(((............(((((((....)))))))......((((....))))...........................................)))............ (-15.70 = -15.70 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:48 2006