
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,760,473 – 16,760,604 |
| Length | 131 |
| Max. P | 0.528465 |

| Location | 16,760,473 – 16,760,567 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 86.97 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -19.71 |
| Energy contribution | -19.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
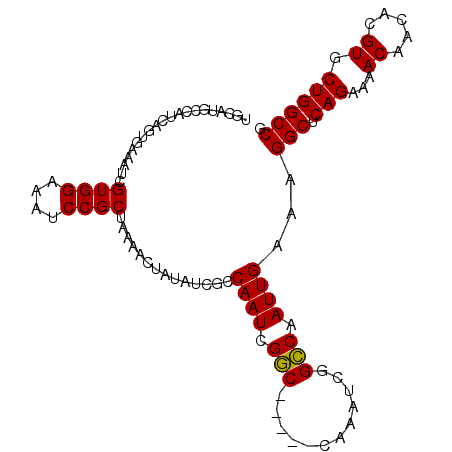
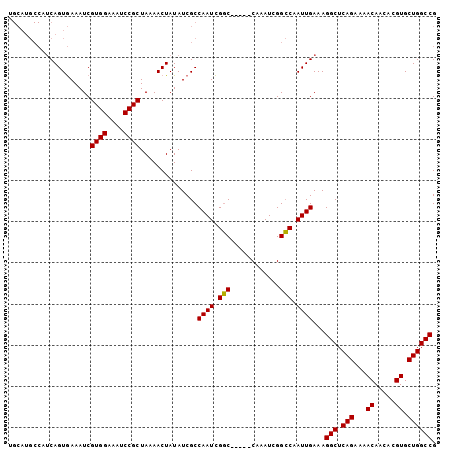
>X_DroMel_CAF1 16760473 94 + 22224390 ----------CAGUAAAAUCGUGGAAAUCCGCUAAAACUAUAUCGCCAAUUGCCAAUCGCCAAUCGGGCAAUUGAAAGGCUCAGAAAACAACACGUGCUGGCCG ----------..........((((....))))..............((((((((............))))))))...(((.(((...((.....)).)))))). ( -24.30) >DroSec_CAF1 47326 99 + 1 UGCAUGCCAUCAGUGAAAUCGUGGAAAUCCGCUAAAACUAUAUCGCCAAUCGGC-----CAAAUCGGCCAAUUGAAAGGCUCAGAAAACAACACGUGCUGGCCG .....((((.(((((.....((((....))))............((((((.(((-----(.....)))).)))....)))...........))).)).)))).. ( -26.30) >DroSim_CAF1 45257 99 + 1 UGCAUGCCAUCAGUGAAAUCGUGGAAAUCCGCUAAAACUAUAUCGCCAAUCGGC-----CAAAUCGGCCAAUUGAAAGGCUCAGAAAACAACACGUGCUGGCCG .....((((.(((((.....((((....))))............((((((.(((-----(.....)))).)))....)))...........))).)).)))).. ( -26.30) >consensus UGCAUGCCAUCAGUGAAAUCGUGGAAAUCCGCUAAAACUAUAUCGCCAAUCGGC_____CAAAUCGGCCAAUUGAAAGGCUCAGAAAACAACACGUGCUGGCCG ....................((((....))))..............((((.(((............))).))))...(((.(((...((.....)).)))))). (-19.71 = -19.27 + -0.44)

| Location | 16,760,491 – 16,760,604 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -28.53 |
| Energy contribution | -27.87 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
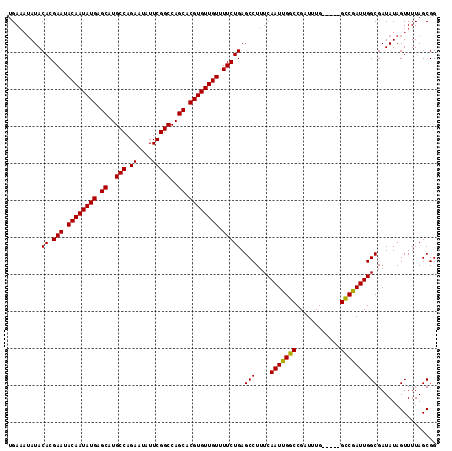
>X_DroMel_CAF1 16760491 113 - 22224390 UGA---AUACACGAAUACAAUAUGAGCAUGCCAGAAUAUUCGGCCAGCACGUGUUGUUUUCUGAGCCUUUCAAUUGCCCGAUUGGCGAUUGGCAAUUGGCGAUAUAGUUUUAGCGG ...---...((.(((.((((((((.((..(((.((....)))))..)).)))))))).))))).(((..(((((((((.....))))))))).....)))................ ( -32.80) >DroSec_CAF1 47354 111 - 1 AGAAAUAUACACGAAUACAAUAUGAGCAUGCCAGAAUAUUCGGCCAGCACGUGUUGUUUUCUGAGCCUUUCAAUUGGCCGAUUUG-----GCCGAUUGGCGAUAUAGUUUUAGCGG .(((((...((.(((.((((((((.((..(((.((....)))))..)).)))))))).))))).(((....((((((((.....)-----))))))))))......)))))..... ( -33.30) >DroSim_CAF1 45285 111 - 1 UGAAAUAUACACGAAUACAAUAUGAGCAUGCCAGAAUAUUCGGCCAGCACGUGUUGUUUUCUGAGCCUUUCAAUUGGCCGAUUUG-----GCCGAUUGGCGAUAUAGUUUUAGCGG ((((((...((.(((.((((((((.((..(((.((....)))))..)).)))))))).))))).(((....((((((((.....)-----))))))))))......)))))).... ( -34.10) >consensus UGAAAUAUACACGAAUACAAUAUGAGCAUGCCAGAAUAUUCGGCCAGCACGUGUUGUUUUCUGAGCCUUUCAAUUGGCCGAUUUG_____GCCGAUUGGCGAUAUAGUUUUAGCGG .........((.(((.((((((((.((..(((.((....)))))..)).)))))))).))))).(((....(((((((............))))))))))................ (-28.53 = -27.87 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:45 2006