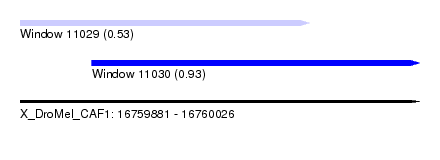
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,759,881 – 16,760,026 |
| Length | 145 |
| Max. P | 0.931770 |

| Location | 16,759,881 – 16,759,986 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.63 |
| Mean single sequence MFE | -17.53 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
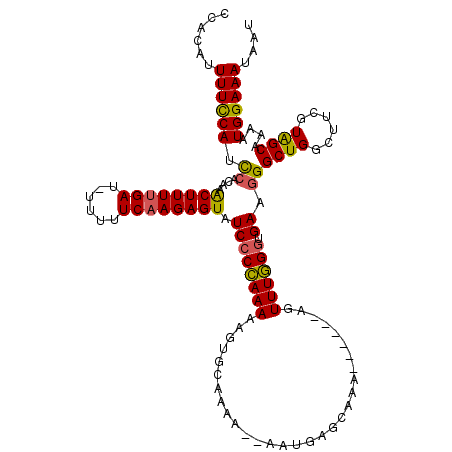
>X_DroMel_CAF1 16759881 105 + 22224390 -----AAAUAGUUGCAAUAAUUUAGCAAAUGCCGAAUUUUCCAUCAACAUACUUUCGAU-UUUUUCAAGAGUAUCGCCAAAAAGUGCAAAAAAUAUGAGCAAACCAGGCAG -----.....((((....((((..((....))..))))......))))(((((((.((.-....)).))))))).(((......(((...........))).....))).. ( -18.70) >DroSec_CAF1 46746 97 + 1 -----AAAUAGAUGCUAUAAAUUAGCAAGUGCCACAUUUUCCAUCCACAAACUUUUGAU-UUUUUCAAGAGUAUCCCCAAAAAGUGCAAAG--UAUGAGCAAA------AU -----.......(((((.....)))))..(((((.(((((.(((......((((((((.-....))))))))...........))).))))--).)).)))..------.. ( -17.23) >DroSim_CAF1 44671 97 + 1 -----AAAUAGAUGCUAUAAACUAGCAAGUGCCACAUUUUCCAACCACAAGCUUUUGAU-UUUUUCAAGAGUAUCCCCAAAAAGUACAAAA--AAUGAGCAAA------AG -----.......(((((.....)))))..(((..((((((..........((((((((.-....)))))))).....(.....).....))--)))).)))..------.. ( -14.00) >DroYak_CAF1 38846 104 + 1 AAGAAAAAUAGGUGCUGUAAAUAAGUAAAUGCAAUAUUUUUCAUUCACAAACUUUUGAAGUAAUUCAAGAGUAUCCCUAAAAAGUGCAAAA--AU---CCAAAAC--UUUG ..((((((((..(((...............))).))))))))........(((((((((....)))))))))........(((((......--..---.....))--))). ( -20.18) >consensus _____AAAUAGAUGCUAUAAAUUAGCAAAUGCCACAUUUUCCAUCCACAAACUUUUGAU_UUUUUCAAGAGUAUCCCCAAAAAGUGCAAAA__AAUGAGCAAA______AG ............(((((.....))))).......................((((((((......))))))))....................................... ( -9.32 = -9.70 + 0.38)



| Location | 16,759,907 – 16,760,026 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.03 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -18.31 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16759907 119 + 22224390 CCGAAUUUUCCAUCAACAUACUUUCGAU-UUUUUCAAGAGUAUCGCCAAAAAGUGCAAAAAAUAUGAGCAAACCAGGCAGUUUGGGUGAAGGGCUGGCUUCGUAGCUAAAUGGAAAUAAU ......(((((((.....((((((.((.-....)).))))))(((((..(((.(((....................))).))).))))).....(((((....))))).))))))).... ( -24.35) >DroSec_CAF1 46772 111 + 1 CCACAUUUUCCAUCCACAAACUUUUGAU-UUUUUCAAGAGUAUCCCCAAAAAGUGCAAAG--UAUGAGCAAA------AUUUUGGGUGAAGGGCUGGCUUCGUAGCAAAAUGGAAAUAAU ......(((((((((....((((((((.-....)))))))).(((((((((..(((....--.....)))..------.))))))).)).))((((......))))...))))))).... ( -30.90) >DroSim_CAF1 44697 111 + 1 CCACAUUUUCCAACCACAAGCUUUUGAU-UUUUUCAAGAGUAUCCCCAAAAAGUACAAAA--AAUGAGCAAA------AGUUUGGGUGAAGGGCUGGCUUUGUAGCAAAAUGGAAACAAU ......((((((.((....((((((((.-....)))))))).((((((((..((.((...--..)).))...------..)))))).)).))((((......))))....)))))).... ( -27.10) >DroYak_CAF1 38877 109 + 1 CAAUAUUUUUCAUUCACAAACUUUUGAAGUAAUUCAAGAGUAUCCCUAAAAAGUGCAAAA--AU---CCAAAAC--UUUGUUUAGGUGAAGGGCUG----UGUGGCAAAAUGGAAAUAAU ......((((((((((((.(((((((((....))))))))).((((((((((((......--..---.....))--))..)))))).)).......----)))))....))))))).... ( -24.32) >consensus CCACAUUUUCCAUCCACAAACUUUUGAU_UUUUUCAAGAGUAUCCCCAAAAAGUGCAAAA__AAUGAGCAAA______AGUUUGGGUGAAGGGCUGGCUUCGUAGCAAAAUGGAAAUAAU ......((((((.((....((((((((......)))))))).((((((((..............................)))))).)).))((((......))))....)))))).... (-18.31 = -17.93 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:42 2006