
| Sequence ID | X_DroMel_CAF1 |
|---|---|
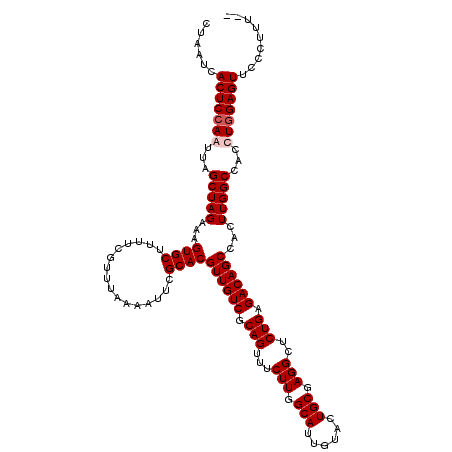
| Location | 16,759,010 – 16,759,195 |
| Length | 185 |
| Max. P | 0.783503 |

| Location | 16,759,010 – 16,759,128 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.76 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -32.31 |
| Energy contribution | -33.07 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16759010 118 + 22224390 CUAAUCACUCCAAUUAGCUAGAAAGUGCUUUUCGUUUAAAAUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCAUUUGGAGUUCCCUUU-- ......(((((((...(((((...((((................))))((((((.(((...(((.(((......))).)))..))).))))))...)))))...))))))).......-- ( -35.49) >DroSec_CAF1 45881 118 + 1 CUAAUCACUCCAAUUAGCUAGAAAGUGCUUUUCGUUUAAAAUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU-- ......((((((....(((((...((((................))))((((((.(((...(((.(((......))).)))..))).))))))...)))))....)))))).......-- ( -33.59) >DroSim_CAF1 43791 118 + 1 CUAAUCACUCGAAUUAGCUAGAAAGUGCUUUUCGUUUAAAAUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU-- ......((((.(....(((((...((((................))))((((((.(((...(((.(((......))).)))..))).))))))...)))))....).)))).......-- ( -29.19) >DroYak_CAF1 37962 120 + 1 CUAAUCACUCCAAUUAGCUAGAAAGUGCUUUUCGUUUAAAAUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCACUUGGAGUUCCCUUUUU ......(((((((...(((((...((((................))))((((((.(((...(((.(((......))).)))..))).))))))...)))))...)))))))......... ( -34.79) >consensus CUAAUCACUCCAAUUAGCUAGAAAGUGCUUUUCGUUUAAAAUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU__ ......(((((((...(((((...((((................))))((((((.(((...(((.(((......))).)))..))).))))))...)))))...)))))))......... (-32.31 = -33.07 + 0.75)

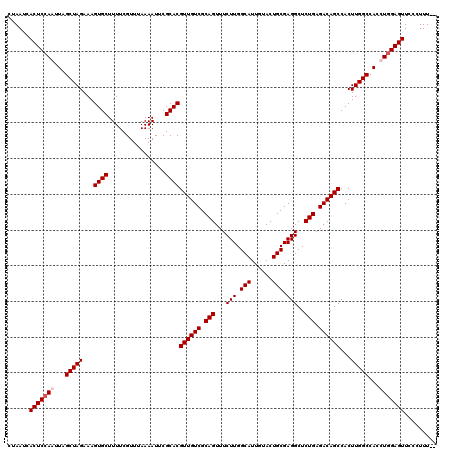
| Location | 16,759,050 – 16,759,159 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -31.99 |
| Energy contribution | -31.80 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16759050 109 + 22224390 AUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCAUUUGGAGUUCCCUUU---AUUCCUUGGCUUGGUCCGAAAAACGAGAAUC-------- .((((...((((((.(((...(((.(((......))).)))..))).)))))).(((.(((((...(((((.......---))))).))))).))).))))...........-------- ( -34.10) >DroSec_CAF1 45921 109 + 1 AUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU---AUUCCUUGGCUUAGUCCGAAAAUCGAGAAUC-------- .((((...((((((.(((...(((.(((......))).)))..))).)))))).(((.(((((...(((((.......---))))).))))).))).))))...........-------- ( -33.40) >DroSim_CAF1 43831 109 + 1 AUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU---AUUCCUUGGCUUAGUCCGAAAAUCGAGAAUC-------- .((((...((((((.(((...(((.(((......))).)))..))).)))))).(((.(((((...(((((.......---))))).))))).))).))))...........-------- ( -33.40) >DroYak_CAF1 38002 120 + 1 AUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCACUUGGAGUUCCCUUUUUAAUUCCUUGGCUCAGUCCUCAAUUCGAGUUCCUACUGCGC .............((((((..(((((((......)))((((..(((((.(.(((....)))..(..((((((........))))))..))))))).))))....)))).....)))))). ( -36.40) >consensus AUUCGCACGUUGUCGCAGUUUCUUGGCAUUGUACUGCGAGGCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU___AUUCCUUGGCUUAGUCCGAAAAUCGAGAAUC________ .((((...((((((.(((...(((.(((......))).)))..))).)))))).(((.(((((...(((((..........))))).))))).)))........))))............ (-31.99 = -31.80 + -0.19)

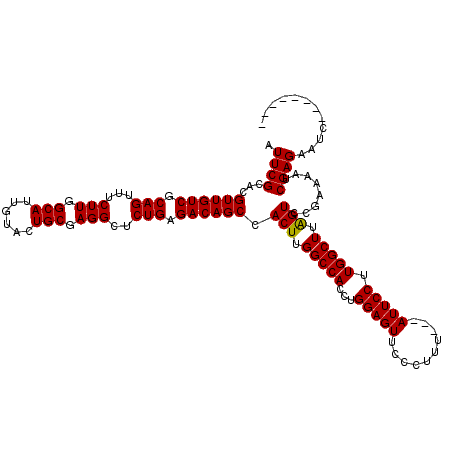

| Location | 16,759,090 – 16,759,195 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16759090 105 + 22224390 GCUCUGAGACAGCCACUUGGCCAUUUGGAGUUCCCUUU---AUUCCUUGGCUUGGUCCGAAAAACGAGAAUC-----------GAUCAUCGUCAAGGCUCAAGGUUCGAGUUUC-AAGUU (((.((((((.((((...(((((...(((((.......---))))).))))))))).((((....(((..(.-----------(((....))).)..)))....)))).)))))-)))). ( -29.70) >DroSec_CAF1 45961 102 + 1 GCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU---AUUCCUUGGCUUAGUCCGAAAAUCGAGAAUC-----------GAU---CGUCAAGGCUCGAGGUUCGAGUUUC-AAGUU (((.((((((.((((((.(((((...(((((.......---))))).))))).)))..((..((((.....)-----------)))---..))..)))(((.....))))))))-)))). ( -33.30) >DroSim_CAF1 43871 102 + 1 GCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU---AUUCCUUGGCUUAGUCCGAAAAUCGAGAAUC-----------GAU---CGUCAAGGCUCGAGGUUCGAGUUUC-AAGUU (((.((((((.((((((.(((((...(((((.......---))))).))))).)))..((..((((.....)-----------)))---..))..)))(((.....))))))))-)))). ( -33.30) >DroYak_CAF1 38042 117 + 1 GCUCUGAGACAGCCACUUGGCCACUUGGAGUUCCCUUUUUAAUUCCUUGGCUCAGUCCUCAAUUCGAGUUCCUACUGCGCCAAGAU---CGCCAAGAUUCAAGGUGCGAGUUUUGAAGUU ((((..((...(((....)))..))..))))...((((..(((((((((((.((((.(((.....))).....)))).))))))..---((((.........)))).)))))..)))).. ( -36.80) >consensus GCUCUGAGACAGCCACUUGGCCACCUGGAGUUCCCUUU___AUUCCUUGGCUUAGUCCGAAAAUCGAGAAUC___________GAU___CGUCAAGGCUCAAGGUUCGAGUUUC_AAGUU (((..(((((....(((.(((((...(((((..........))))).))))).))).((((..(((((.............................)))))..)))).)))))..))). (-21.71 = -21.77 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:37 2006