
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,757,457 – 16,757,636 |
| Length | 179 |
| Max. P | 0.997198 |

| Location | 16,757,457 – 16,757,575 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -28.17 |
| Energy contribution | -28.43 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16757457 118 - 22224390 GCAGUUGUGUCAACAAAAGUAUGAUGCCCGCGUUGAAGGGUAUCGCACUUGACACAUGCCUGGGUUUAUUCACAC--ACCGAUCGGCCAUAAGUCGGUUAUGAAGCUCGGAGCUUGGAGC (((..(((((((.....(((.((((((((........)))))))).)))))))))))))(..(((((.((((...--((((((.........))))))..)))).....)))))..)... ( -40.20) >DroSec_CAF1 44438 105 - 1 GCAGUUGUGUCAACAAAAGUAUGAUGCCCGCGUUGAAGGGUAUCGCACUUGACACAUGUCUGGGUUUAUUCACAC--ACCGAUCGGCCAUAAGUCGGCUAUGAGUCU------------- .(((.(((((((.....(((.((((((((........)))))))).))))))))))...))).....(((((...--.(((((.........)))))...)))))..------------- ( -32.20) >DroEre_CAF1 35779 105 - 1 GCAGUUGUGUCAACAA-AGUAUGAUGCCCGCGUCGAAGGGUAUCGCACUUGACACAUGCCUGGGUUUAUUCACACACACUGAUCGGCCAUAAGUCGGCUGUGAAGC-------------- .(((.(((((((....-(((.((((((((........)))))))).))))))))))...)))......(((((.(.....)...((((.......)))))))))..-------------- ( -33.70) >DroYak_CAF1 36242 93 - 1 GCAGUUGUGUCAACAAAAGUAUGAUGCCCGCGUUGAAGGGUAUCGCACUUGACACAUGCCUGGGUUUAUUCACAC--ACCGAUCGG-----------CUGUGAAGC-------------- .(((.(((((((.....(((.((((((((........)))))))).))))))))))...)))......((((((.--.((....))-----------.))))))..-------------- ( -32.30) >consensus GCAGUUGUGUCAACAAAAGUAUGAUGCCCGCGUUGAAGGGUAUCGCACUUGACACAUGCCUGGGUUUAUUCACAC__ACCGAUCGGCCAUAAGUCGGCUAUGAAGC______________ .(((.(((((((.....(((.((((((((........)))))))).))))))))))...)))......(((((...........((((.......)))))))))................ (-28.17 = -28.43 + 0.25)

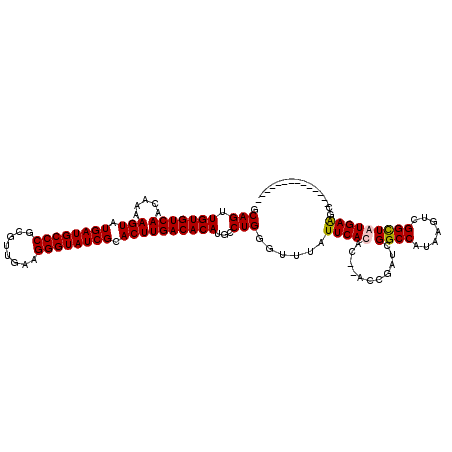
| Location | 16,757,497 – 16,757,611 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -41.25 |
| Consensus MFE | -30.95 |
| Energy contribution | -35.08 |
| Covariance contribution | 4.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16757497 114 - 22224390 AAGCGAGUGUAUCACAGGCACCA----UUGGCUGUGCGCAGCAGUUGUGUCAACAAAAGUAUGAUGCCCGCGUUGAAGGGUAUCGCACUUGACACAUGCCUGGGUUUAUUCACAC--ACC ..(.(((((.(((.((((((...----...((((....))))...(((((((.....(((.((((((((........)))))))).))))))))))))))))))).))))).)..--... ( -45.60) >DroSec_CAF1 44465 118 - 1 AAGCGAGUGUAUCACAGGCACCGUCAACUGGCUACGCGCAGCAGUUGUGUCAACAAAAGUAUGAUGCCCGCGUUGAAGGGUAUCGCACUUGACACAUGUCUGGGUUUAUUCACAC--ACC ..(.(((((.(((.((((((.....(((((.((......)))))))((((((.....(((.((((((((........)))))))).))))))))).))))))))).))))).)..--... ( -40.70) >DroEre_CAF1 35805 119 - 1 AAGCGACUGCAUCACAGGCACCGCCAACUGGCUGUGCACAGCAGUUGUGUCAACAA-AGUAUGAUGCCCGCGUCGAAGGGUAUCGCACUUGACACAUGCCUGGGUUUAUUCACACACACU ..(.((.((.(((.((((((.....(((((.(((....))))))))((((((....-(((.((((((((........)))))))).))))))))).))))))))).)).)).)....... ( -43.60) >DroYak_CAF1 36257 91 - 1 AAGCGA---------------------------GUGUGCCGCAGUUGUGUCAACAAAAGUAUGAUGCCCGCGUUGAAGGGUAUCGCACUUGACACAUGCCUGGGUUUAUUCACAC--ACC ..(.((---------------------------(((.(((.(((.(((((((.....(((.((((((((........)))))))).))))))))))...)))))).))))).)..--... ( -35.10) >consensus AAGCGAGUGUAUCACAGGCACCG____CUGGCUGUGCGCAGCAGUUGUGUCAACAAAAGUAUGAUGCCCGCGUUGAAGGGUAUCGCACUUGACACAUGCCUGGGUUUAUUCACAC__ACC ..(.(((((.(((.((((((..........((((....))))...(((((((.....(((.((((((((........)))))))).))))))))))))))))))).))))).)....... (-30.95 = -35.08 + 4.12)


| Location | 16,757,535 – 16,757,636 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -22.85 |
| Energy contribution | -22.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16757535 101 + 22224390 CCCUUCAACGCGGGCAUCAUACUUUUGUUGACACAACUGCUGCGCACAGCCAA----UGGUGCCUGUGAUACACUCGCUUUGCGACUCGCUUUUCCAAUUAUUUU ........((((((((((((......((((...)))).((((....))))..)----)))))))))))......((((...)))).................... ( -27.00) >DroSec_CAF1 44503 100 + 1 CCCUUCAACGCGGGCAUCAUACUUUUGUUGACACAACUGCUGCGCGUAGCCAGUUGACGGUGCCUGUGAUACACUCGCUUUGC-----GCAUUUCCAAUUAUUUU ........((((((((((.......((....))(((((((((....))).))))))..))))))))))........((.....-----))............... ( -27.50) >DroEre_CAF1 35845 99 + 1 CCCUUCGACGCGGGCAUCAUACU-UUGUUGACACAACUGCUGUGCACAGCCAGUUGGCGGUGCCUGUGAUGCAGUCGCUUUGU-----GGGUUUCCCAUUCUUUU (((.....((((((((((.....-.((....))(((((((((....))).))))))..))))))))))..((((.....))))-----))).............. ( -32.90) >consensus CCCUUCAACGCGGGCAUCAUACUUUUGUUGACACAACUGCUGCGCACAGCCAGUUG_CGGUGCCUGUGAUACACUCGCUUUGC_____GCAUUUCCAAUUAUUUU ........((((((((((........((((...)))).((((....))))........))))))))))..................................... (-22.85 = -22.63 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:34 2006