
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,757,205 – 16,757,417 |
| Length | 212 |
| Max. P | 0.981305 |

| Location | 16,757,205 – 16,757,325 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.06 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -22.75 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16757205 120 + 22224390 UUAUUCAAAUGCCCACCAAUGCCAAUCGAAUGUAUGUAUGGUUUUUUUUUUUUGCUCCACCUAAGUCUUGGCAUUUUUUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGC .................((((((((..((...((.((..(((...........)))..)).))..))))))))))..((((((((......(((((((.....))))))).)))))))). ( -25.40) >DroSec_CAF1 44201 113 + 1 UUAUUCAAAUGCCCACCAAUGCCAAUCGAA---GUGUAUGGUUUUUUUU----UCUCCACCUAAGUCUUGGCAUUUUUUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGC .................((((((((..((.---.((..(((........----...)))..))..))))))))))..((((((((......(((((((.....))))))).)))))))). ( -27.00) >DroEre_CAF1 35576 108 + 1 UUAUUCAAAUGCCCGCCAAUGCCAAUCGAA---GUGUAUUGUUUUUCU--------CCACCCAAGUCUUGGCAUUU-UUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGC .................((((((((..((.---.((...((.......--------.))..))..)))))))))).-((((((((......(((((((.....))))))).)))))))). ( -24.50) >DroYak_CAF1 35999 116 + 1 UUAUUCAAAUGCCCACCAAUGCCAAUCGAA---GUGUAUUGUUUUUUUU-UUUGGUUCACCCAAGUCUUGGCAUUUUUUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGC ......(((((((...(((((((.......---).))))))........-(((((.....)))))....))))))).((((((((......(((((((.....))))))).)))))))). ( -29.40) >consensus UUAUUCAAAUGCCCACCAAUGCCAAUCGAA___GUGUAUGGUUUUUUUU____GCUCCACCCAAGUCUUGGCAUUUUUUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGC .................((((((((..((....(((.....................))).....))))))))))..((((((((......(((((((.....))))))).)))))))). (-22.75 = -23.00 + 0.25)



| Location | 16,757,205 – 16,757,325 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.06 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.21 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16757205 120 - 22224390 GCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAAUAAAAAAUGCCAAGACUUAGGUGGAGCAAAAAAAAAAAACCAUACAUACAUUCGAUUGGCAUUGGUGGGCAUUUGAAUAA ..((((..((((.(((.(((((......))))).((((........(((((((.......((((...............))))............))))))))))))))))))..)))). ( -27.27) >DroSec_CAF1 44201 113 - 1 GCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAAUAAAAAAUGCCAAGACUUAGGUGGAGA----AAAAAAAACCAUACAC---UUCGAUUGGCAUUGGUGGGCAUUUGAAUAA ..((((..((((.(((.(((((......))))).((((........(((((((.....(((((....----.............)))---))...))))))))))))))))))..)))). ( -29.93) >DroEre_CAF1 35576 108 - 1 GCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAAUAA-AAAUGCCAAGACUUGGGUGG--------AGAAAAACAAUACAC---UUCGAUUGGCAUUGGCGGGCAUUUGAAUAA ..((((..((((.(((.(((((......)))))..........-.((((((((.....(((((.--------............)))---))...))))))))...)))))))..)))). ( -29.42) >DroYak_CAF1 35999 116 - 1 GCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAAUAAAAAAUGCCAAGACUUGGGUGAACCAAA-AAAAAAAACAAUACAC---UUCGAUUGGCAUUGGUGGGCAUUUGAAUAA ..((((..((((.(((.(((((......))))).((((........(((((((.....(((((.......-.............)))---))...))))))))))))))))))..)))). ( -28.35) >consensus GCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAAUAAAAAAUGCCAAGACUUAGGUGGAGC____AAAAAAAACAAUACAC___UUCGAUUGGCAUUGGUGGGCAUUUGAAUAA ..((((..((((.(((.(((((......)))))............((((((((.......(((.....................)))........))))))))...)))))))..)))). (-26.40 = -26.21 + -0.19)



| Location | 16,757,245 – 16,757,351 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16757245 106 + 22224390 GUUUUUUUUUUUUGCUCCACCUAAGUCUUGGCAUUUUUUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCAAAUAUUUUGUGGGGCUCU------UGAUCGU-------- .............(((((((....((....))((((.((((((((......(((((((.....))))))).)))))))).)))).....)))))))...------.......-------- ( -29.40) >DroSec_CAF1 44238 102 + 1 GUUUUUUUU----UCUCCACCUAAGUCUUGGCAUUUUUUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCAAAUAUUUUGUGCGGCUCU------UGAUCGU-------- .........----........((((...(.((((....((((((.(((...(((((((.....)))))))..)))....))))))....)))).)..))------)).....-------- ( -20.80) >DroEre_CAF1 35613 110 + 1 GUUUUUCU--------CCACCCAAGUCUUGGCAUUU-UUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCA-AUGCUUUGUGGGGCUUCAAGUCGCGAUCGCAGCUCUCA ........--------...((((......((((((.-((((((((......(((((((.....))))))).)))))))).)-)))))...))))......(((.((....)).))).... ( -33.30) >DroYak_CAF1 36036 119 + 1 GUUUUUUUU-UUUGGUUCACCCAAGUCUUGGCAUUUUUUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCAAAUAUUUUGUGGGGCUCCAAGUCGCGAUCGCAGCUCUUA .........-.((((..(.((((.((....))((((.((((((((......(((((((.....))))))).)))))))).))))......)))))..))))((.((....)).))..... ( -30.60) >consensus GUUUUUUUU____GCUCCACCCAAGUCUUGGCAUUUUUUAUUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCAAAUAUUUUGUGGGGCUCC______CGAUCGC________ ...................((((.((....))((((.((((((((......(((((((.....))))))).)))))))).))))......)))).......................... (-20.60 = -20.85 + 0.25)



| Location | 16,757,285 – 16,757,383 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -22.35 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
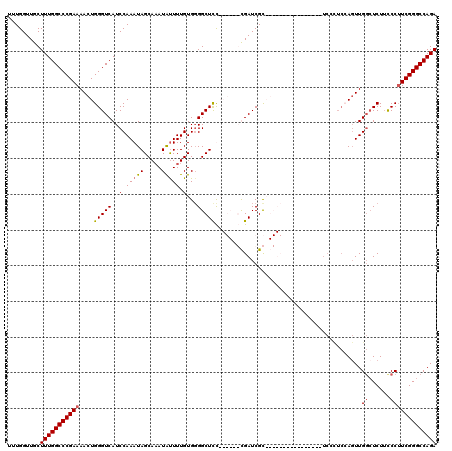
>X_DroMel_CAF1 16757285 98 + 22224390 UUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCAAAUAUUUUGUGGGGCUCU------UGAUCGU----------------UCACUCCAGUUGGCUCUUCCCAUCGGGCCAGA .........((((((((((....((((....((((...((((.....))))((((....------(((....----------------)))))))..)))).....)))))))))))))) ( -31.00) >DroSec_CAF1 44274 98 + 1 UUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCAAAUAUUUUGUGCGGCUCU------UGAUCGU----------------UCCUUCCAGUUGGGUCUUCCCUUCGGGCCAGA .........((((((((((((((((((...........((((.....))))(((((...------.).))))----------------....)))))))(((....)))))))))))))) ( -32.40) >DroEre_CAF1 35644 103 + 1 UUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCA-AUGCUUUGUGGGGCUUCAAGUCGCGAUCGCAGCUCUCA---------------ACUUGCCUCU-UCCUUCGGGCCAGA .........(((((((((((.....((((..(((...(((.-..)))...)))(((.....)))..))))((((......---------------..))))....-...))))))))))) ( -31.40) >DroYak_CAF1 36075 120 + 1 UUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCAAAUAUUUUGUGGGGCUCCAAGUCGCGAUCGCAGCUCUUAAACACAACUCCCUCGAGUUGGCUCUUUCCUUCGGGCCAGA .........(((((((((((..(((((((.........((((.....))))(.(((.....))).))))).)))..........((((((....)))))).........))))))))))) ( -36.70) >consensus UUUGGUUGCUUUGGCCCGAAAACUGGGUCAUCCAAAUAGCAAAUAUUUUGUGGGGCUCC______CGAUCGC________________UCCCUCCAGUUGGCUCUUCCCUUCGGGCCAGA .........(((((((((((....(((((..(.(((((.....))))).)...))))).........................................((......))))))))))))) (-22.35 = -22.73 + 0.37)


| Location | 16,757,285 – 16,757,383 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.34 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
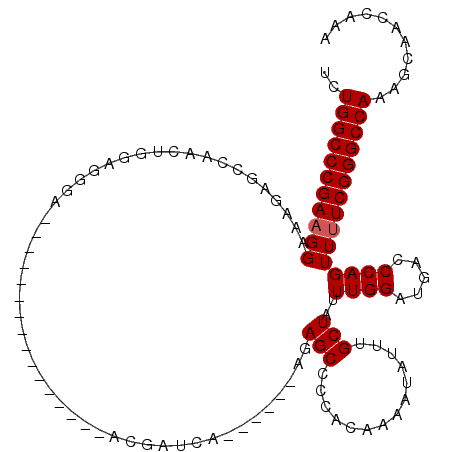
>X_DroMel_CAF1 16757285 98 - 22224390 UCUGGCCCGAUGGGAAGAGCCAACUGGAGUGA----------------ACGAUCA------AGAGCCCCACAAAAUAUUUGCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAA ..(((((((((((......)))(((((.((..----------------.((.(((------(((((..............))).))))).)))))))))..))))))))........... ( -27.64) >DroSec_CAF1 44274 98 - 1 UCUGGCCCGAAGGGAAGACCCAACUGGAAGGA----------------ACGAUCA------AGAGCCGCACAAAAUAUUUGCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAA ..((((((((((((....)))((((((..((.----------------.(.....------.)..))(((.........)))............)))))))))))))))........... ( -31.40) >DroEre_CAF1 35644 103 - 1 UCUGGCCCGAAGGA-AGAGGCAAGU---------------UGAGAGCUGCGAUCGCGACUUGAAGCCCCACAAAGCAU-UGCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAA ..(((((((((((.-.(.(((((((---------------((.((.......)).))))))).....(((.(.(((..-.))).).)))....)))..)))))))))))........... ( -33.10) >DroYak_CAF1 36075 120 - 1 UCUGGCCCGAAGGAAAGAGCCAACUCGAGGGAGUUGUGUUUAAGAGCUGCGAUCGCGACUUGGAGCCCCACAAAAUAUUUGCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAA ..(((((((((((...((((((((((....)))))).))))....(((.(((.......))).))).))................((((.....))))..)))))))))........... ( -36.30) >consensus UCUGGCCCGAAGGAAAGAGCCAACUGGAGGGA________________ACGAUCA______AGAGCCCCACAAAAUAUUUGCUAUUUGGAUGACCCAGUUUUCGGGCCAAAGCAACCAAA ..(((((((((((..................................................(((..............)))..((((.....)))))))))))))))........... (-20.09 = -20.34 + 0.25)


| Location | 16,757,325 – 16,757,417 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.24 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -19.47 |
| Energy contribution | -19.81 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
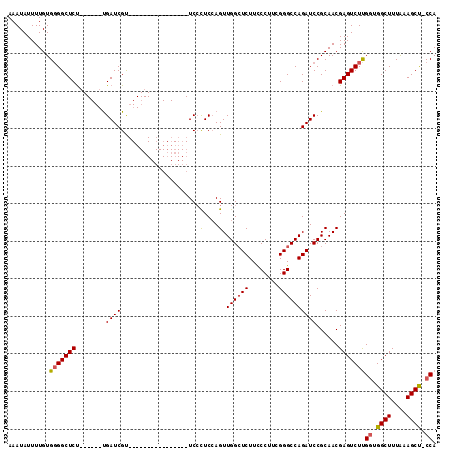
>X_DroMel_CAF1 16757325 92 + 22224390 AAAUAUUUUGUGGGGCUCU------UGAUCGU----------------UCACUCCAGUUGGCUCUUCCCAUCGGGCCAGAUCCGCAACGAGUCUUGGUGGCUUUAUAGCCACCA ...........(((((((.------(((....----------------)))......(((((((........))))))).........)))))))(((((((....))))))). ( -31.20) >DroSec_CAF1 44314 91 + 1 AAAUAUUUUGUGCGGCUCU------UGAUCGU----------------UCCUUCCAGUUGGGUCUUCCCUUCGGGCCAGAUCCGCAACGAGUCUUGGUGGCUUUGCAGCU-CCA ..........((.((((..------.......----------------...........((((((..((....))..))))))(((..(((((.....))))))))))))-.)) ( -24.10) >DroYak_CAF1 36115 113 + 1 AAAUAUUUUGUGGGGCUCCAAGUCGCGAUCGCAGCUCUUAAACACAACUCCCUCGAGUUGGCUCUUUCCUUCGGGCCAGAUCCGCAACGAGUCUCGCCGGCUUUAAAGCU-CCA ..........(((((((..((((((((((((..((..........(((((....)))))((.(((..((....))..))).))))..)))...))).))))))...))))-))) ( -36.50) >consensus AAAUAUUUUGUGGGGCUCU______UGAUCGU________________UCCCUCCAGUUGGCUCUUCCCUUCGGGCCAGAUCCGCAACGAGUCUUGGUGGCUUUAAAGCU_CCA ...........(((((((........((((.....................(....).((((((........))))))))))......)))))))((.((((....)))).)). (-19.47 = -19.81 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:31 2006