
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,754,109 – 16,754,399 |
| Length | 290 |
| Max. P | 0.998331 |

| Location | 16,754,109 – 16,754,215 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.37 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.793936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16754109 106 - 22224390 CAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG------------AGAAAUAAAAGCAC--ACAAAAAACCCAACAAAUCAUCGCGUAAUACAG ..(((((...((((((((((......(((((((...(((((......))))).))..))------------)))...))))))))--))....................)))))...... ( -22.80) >DroSec_CAF1 41151 106 - 1 CAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG------------AGAAAUAAAAGCAC--ACAAAAAAGCCAACAAAUCACCGCGUAAUACAG ..(((((...((((((((((......(((((((...(((((......))))).))..))------------)))...))))))))--))......(....)........)))))...... ( -25.10) >DroSim_CAF1 38314 106 - 1 CAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG------------AGAAAUAAAAGCAC--ACAAAAAACCCAACAAAUCACCGCGUAAUACAG ..(((((...((((((((((......(((((((...(((((......))))).))..))------------)))...))))))))--))....................)))))...... ( -22.80) >DroEre_CAF1 32663 105 - 1 CAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG-------------AAAAUAAAAGCAC--ACAAAAAACCCAACAAAUCACCGCGUAAUACAG ..(((((.((((((((.....)))))..........(((((......)))))))).((.-------------.........))..--......................)))))...... ( -21.10) >DroYak_CAF1 32963 120 - 1 CAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCGCAGUGCAGCGCGAGAAAUAAAAGCACACACAAAAAAGCCAACAAAUCACCGCGUAAUACAG ..(((((.(.((((((((((......((((.((...(((((......))))).)).((((......))))))))...)))))))))).)......(....)........)))))...... ( -31.70) >consensus CAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG____________AGAAAUAAAAGCAC__ACAAAAAACCCAACAAAUCACCGCGUAAUACAG ..(((((.((((((((.....)))))..........(((((......)))))))).((.......................))..........................)))))...... (-20.20 = -20.20 + -0.00)


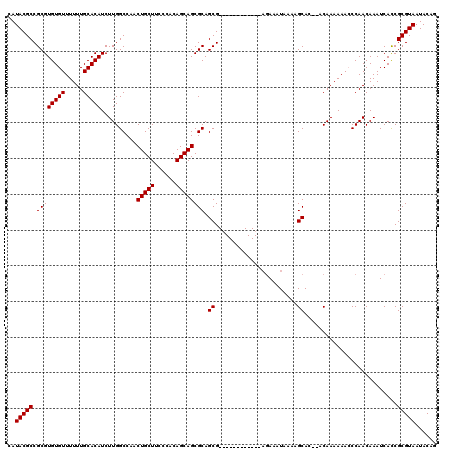
| Location | 16,754,147 – 16,754,255 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -33.70 |
| Energy contribution | -34.30 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16754147 108 + 22224390 UUUAUUUCU------------CGCUGCGCUGCUGUGGGAAGCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGCGCAAUGUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUG .........------------.(((((((((((......)))))).........((((.......)))).)))))(((((((((.(((((((....)))....)))).)))))))))... ( -38.20) >DroSec_CAF1 41189 108 + 1 UUUAUUUCU------------CGCUGCGCUGCUGUGGGAAGCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGCGCAAUAUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUG .........------------.(((((((((((......)))))).........((((.......)))).)))))(((((((((...(((((....)))))((...)))))))))))... ( -37.40) >DroSim_CAF1 38352 108 + 1 UUUAUUUCU------------CGCUGCGCUGCUGUGGGAAGCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGCGCAAUAUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUG .........------------.(((((((((((......)))))).........((((.......)))).)))))(((((((((...(((((....)))))((...)))))))))))... ( -37.40) >DroEre_CAF1 32701 106 + 1 UUUAUUUU-------------CGCUGCGCUGCUGUGGGAAGCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGAGCAAUGC-UGCCCGGGCAGAGCAACGCUGCGUAUACUUG ........-------------.((((.((((((......))))))))))((((..(((.......)))(((((((((....((....(-(((....)))).)).)))))))))...)))) ( -36.90) >DroYak_CAF1 33003 120 + 1 UUUAUUUCUCGCGCUGCACUGCGCUGCGCUGCUGUGGGAAGCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGAGCAAUGCUUGCCCGGGCAGAGCAACGCUGCGUAUACGUG .........((((.(((((((.((((.((((((......))))))))))))....)))))........(((((((((....((..(((((....)))))..)).)))))))))...)))) ( -42.80) >consensus UUUAUUUCU____________CGCUGCGCUGCUGUGGGAAGCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGCGCAAUGUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUG ......................(((((((((((......)))))).........((((.......)))).)))))((((((((.....((((....))))(((...)))))))))))... (-33.70 = -34.30 + 0.60)

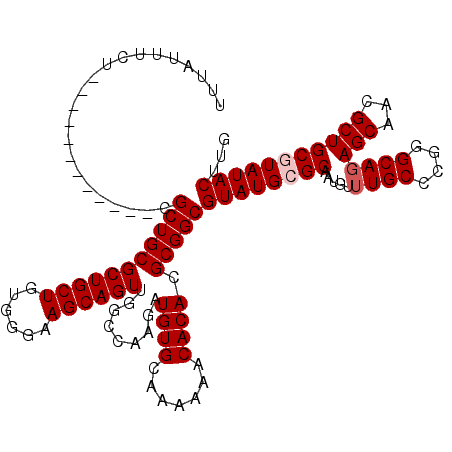
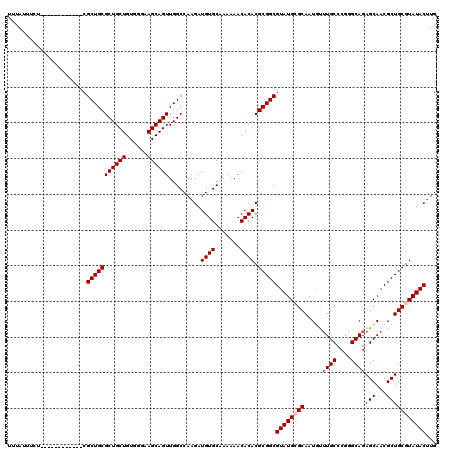
| Location | 16,754,147 – 16,754,255 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -37.64 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16754147 108 - 22224390 CAAGUAUACGCAGCGUUGCUCUGCCCGGGCAAACAUUGCGCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG------------AGAAAUAAA ........(((.((((((((.((...(((((.....)))(((...((((.((((((.....))))))..))))....)))..)))).)))))))).)))------------......... ( -35.50) >DroSec_CAF1 41189 108 - 1 CAAGUAUACGCAGCGUUGCUCUGCCCGGGCAAAUAUUGCGCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG------------AGAAAUAAA ........(((.((((((((.((...(((((.....)))(((...((((.((((((.....))))))..))))....)))..)))).)))))))).)))------------......... ( -35.50) >DroSim_CAF1 38352 108 - 1 CAAGUAUACGCAGCGUUGCUCUGCCCGGGCAAAUAUUGCGCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG------------AGAAAUAAA ........(((.((((((((.((...(((((.....)))(((...((((.((((((.....))))))..))))....)))..)))).)))))))).)))------------......... ( -35.50) >DroEre_CAF1 32701 106 - 1 CAAGUAUACGCAGCGUUGCUCUGCCCGGGCA-GCAUUGCUCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG-------------AAAAUAAA ........(((.((((((((......(((.(-((((((.......((((.((((((.....))))))..))))))).)))).)))..)))))))).)))-------------........ ( -36.51) >DroYak_CAF1 33003 120 - 1 CACGUAUACGCAGCGUUGCUCUGCCCGGGCAAGCAUUGCUCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCGCAGUGCAGCGCGAGAAAUAAA (.(((...(((.(((((((.((((..(((.((((((((.......((((.((((((.....))))))..))))))).))))))))...)))).))))))).)))..))).)......... ( -45.21) >consensus CAAGUAUACGCAGCGUUGCUCUGCCCGGGCAAACAUUGCGCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGCUUCCCACAGCAGCGCAGCG____________AGAAAUAAA ........(((.((((((((.((...(.(((.....))).)....((((.((((((.....))))))..))))...........)).)))))))).)))..................... (-32.28 = -32.28 + 0.00)

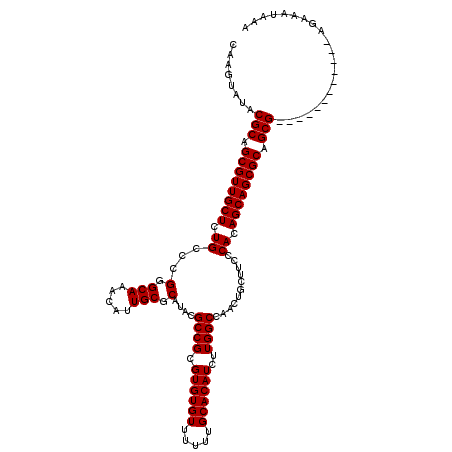

| Location | 16,754,175 – 16,754,292 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -31.02 |
| Energy contribution | -31.94 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16754175 117 + 22224390 GCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGCGCAAUGUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGCAAAGAGAA--U-UACGUCUCUUCUCUUGCAGAAUGU ((((..(((((((..(((.......)))(((((((((.(((......(((((....)))))))))))))))))...))))..)).((((((.--.-....)))))))..))))....... ( -36.40) >DroSec_CAF1 41217 115 + 1 GCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGCGCAAUAUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGCAAAGAGAA--A-UACAU--CUUCUCUUGCAGAAUGU (((......((((..(((.......)))(((((((((.(((......(((((....)))))))))))))))))...)))).(((((.(((((--.-.....--.))))))))))...))) ( -37.00) >DroSim_CAF1 38380 115 + 1 GCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGCGCAAUAUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGCAAAGAGAA--A-UACAU--CUUCUCUUGCAGAAUGC (((......((((..(((.......)))(((((((((.(((......(((((....)))))))))))))))))...)))).(((((.(((((--.-.....--.))))))))))...))) ( -38.40) >DroEre_CAF1 32728 117 + 1 GCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGAGCAAUGC-UGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGUGAAGAGAAAAGCGACGA--UUUCUCUUGCAGAAUGU (((....((((((..(((.......)))(((((((((....((....(-(((....)))).)).)))))))))...)))).....((((((((........--))))))))))....))) ( -36.70) >DroYak_CAF1 33043 117 + 1 GCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGAGCAAUGCUUGCCCGGGCAGAGCAACGCUGCGUAUACGUGAUGCAAAAAGAGAAA-UACGA--UUUCUCUUGCAGAAUGU ...(((.(((.....).))....)))..(((((((((....((..(((((....)))))..)).)))))))))..((((..((((...(((((((-(...)--)))))))))))..)))) ( -37.90) >consensus GCAGUUGGCCAAGAUGUGCAAAAAACACACGCGGCGUAUGCGCAAUGUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGCAAAGAGAA__A_UACGU__CUUCUCUUGCAGAAUGU (((......((((..(((.......)))(((((((((.(((......(((((....)))))))))))))))))...)))).(((.(((((((............))))))))))...))) (-31.02 = -31.94 + 0.92)



| Location | 16,754,175 – 16,754,292 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -32.46 |
| Energy contribution | -32.62 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16754175 117 - 22224390 ACAUUCUGCAAGAGAAGAGACGUA-A--UUCUCUUUGCAUCAAGUAUACGCAGCGUUGCUCUGCCCGGGCAAACAUUGCGCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGC ......((((((((((........-.--))))).)))))....((((.(((((..((((((.....))))))...))))).))))((((.((((((.....))))))..))))....... ( -37.60) >DroSec_CAF1 41217 115 - 1 ACAUUCUGCAAGAGAAG--AUGUA-U--UUCUCUUUGCAUCAAGUAUACGCAGCGUUGCUCUGCCCGGGCAAAUAUUGCGCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGC ......(((((((((((--.....-)--))))).)))))....((((.(((((..((((((.....))))))...))))).))))((((.((((((.....))))))..))))....... ( -37.70) >DroSim_CAF1 38380 115 - 1 GCAUUCUGCAAGAGAAG--AUGUA-U--UUCUCUUUGCAUCAAGUAUACGCAGCGUUGCUCUGCCCGGGCAAAUAUUGCGCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGC (((...(((((((((((--.....-)--))))).)))))....((((.(((((..((((((.....))))))...))))).))))((((.((((((.....))))))..))))....))) ( -38.40) >DroEre_CAF1 32728 117 - 1 ACAUUCUGCAAGAGAAA--UCGUCGCUUUUCUCUUCACAUCAAGUAUACGCAGCGUUGCUCUGCCCGGGCA-GCAUUGCUCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGC ......((.((((((((--........))))))))))......((((..((((.(((((((.....)))))-)).))))..))))((((.((((((.....))))))..))))....... ( -36.40) >DroYak_CAF1 33043 117 - 1 ACAUUCUGCAAGAGAAA--UCGUA-UUUCUCUUUUUGCAUCACGUAUACGCAGCGUUGCUCUGCCCGGGCAAGCAUUGCUCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGC ......(((((((((((--(...)-)))))))...))))....((((..(.((((.((((.(((....))))))).)))))))))((((.((((((.....))))))..))))....... ( -34.90) >consensus ACAUUCUGCAAGAGAAG__ACGUA_U__UUCUCUUUGCAUCAAGUAUACGCAGCGUUGCUCUGCCCGGGCAAACAUUGCGCAUACGCCGCGUGUGUUUUUUGCACAUCUUGGCCAACUGC ......((((((((((............))))))).)))....((((.(((((.(((((((.....))))).)).))))).))))((((.((((((.....))))))..))))....... (-32.46 = -32.62 + 0.16)

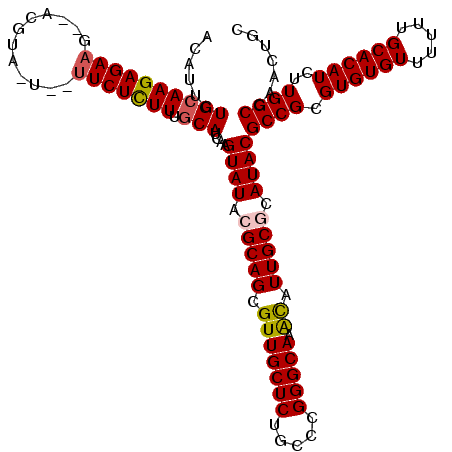

| Location | 16,754,215 – 16,754,332 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16754215 117 + 22224390 CGCAAUGUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGCAAAGAGAA--U-UACGUCUCUUCUCUUGCAGAAUGUUUCAGGUUGCAAAUAGCUUUGGUUGCAAGAGCUAGCCAUU .(((.....)))...(((...(((((.(((.....((....(((((.(((((--.-........))))))))))....))..))))))))...(((((((.......))))))))))... ( -33.90) >DroSec_CAF1 41257 115 + 1 CGCAAUAUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGCAAAGAGAA--A-UACAU--CUUCUCUUGCAGAAUGUUUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCCAGCCAUU .((....(((((....)))))(((((.(((.....((....(((((.(((((--.-.....--.))))))))))....))..)))))))).....))...((((((....).)))))... ( -34.20) >DroSim_CAF1 38420 115 + 1 CGCAAUAUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGCAAAGAGAA--A-UACAU--CUUCUCUUGCAGAAUGCUUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCCAGCCAUU .((....(((((....)))))(((((.(((.((((......(((((.(((((--.-.....--.))))))))))..))))..)))))))).....))...((((((....).)))))... ( -34.70) >DroEre_CAF1 32768 117 + 1 AGCAAUGC-UGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGUGAAGAGAAAAGCGACGA--UUUCUCUUGCAGAAUGUUUCAGGUUGCACAUAGCUAUGGUUGCAAGUGCUAGCGCUU .(((((.(-(((....)))).(((((.(((.....((....(((.((((((((........--)))))))))))....))..))))))))...........)))))(((((....))))) ( -35.10) >DroYak_CAF1 33083 117 + 1 AGCAAUGCUUGCCCGGGCAGAGCAACGCUGCGUAUACGUGAUGCAAAAAGAGAAA-UACGA--UUUCUCUUGCAGAAUGUUGCAGGUUGCAAAUAGCAUCGGUUGCAAGAGCUAGCAAUU (((....(((((((((((...(((((.(((((...((((..((((...(((((((-(...)--)))))))))))..)))))))))))))).....)).))))..))))).)))....... ( -41.30) >consensus CGCAAUGUUUGCCCGGGCAGAGCAACGCUGCGUAUACUUGAUGCAAAGAGAA__A_UACGU__CUUCUCUUGCAGAAUGUUUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCUAGCCAUU .(((((.(((((....)))))(((((.(((.....((....(((.(((((((............))))))))))....))..))))))))...........))))).............. (-26.82 = -27.50 + 0.68)

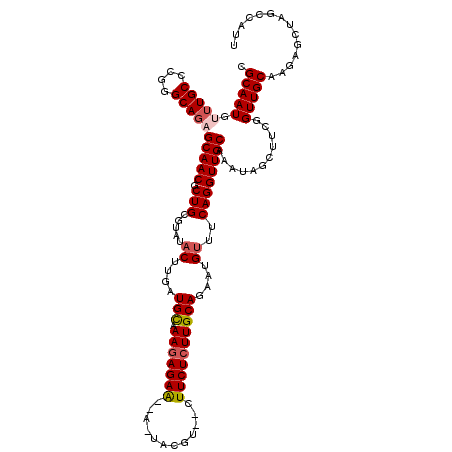
| Location | 16,754,255 – 16,754,365 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.28 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16754255 110 + 22224390 AUGCAAAGAGAA--U-UACGUCUCUUCUCUUGCAGAAUGUUUCAGGUUGCAAAUAGCUUUGGUUGCAAGAGCUAGCCAUUCGAAAA---UGCAC--GAGUUGU--GUUCUUGAAAUUGAA .(((((.(((((--.-........))))))))))....((((((((..((...(((((((.......)))))))((.(((((....---....)--)))).))--)).)))))))).... ( -28.00) >DroSec_CAF1 41297 110 + 1 AUGCAAAGAGAA--A-UACAU--CUUCUCUUGCAGAAUGUUUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCCAGCCAUUCGAAAA---UGCAAUGCAGUUGU--GUUCUUGAAAUUGAA ..((.(((((((--.-.....--.)))))))((((..((...))..)))).....)).((((((.((((((((((((((((.....---.).))))..)))).--))))))).)))))). ( -28.60) >DroSim_CAF1 38460 110 + 1 AUGCAAAGAGAA--A-UACAU--CUUCUCUUGCAGAAUGCUUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCCAGCCAUUCGAAAA---UGCAAUGCAGUUGU--GUUCUUGAAAUUGAA .(((((.(((((--.-.....--.))))))))))...(((........))).......((((((.((((((((((((((((.....---.).))))..)))).--))))))).)))))). ( -29.20) >DroEre_CAF1 32807 111 + 1 AUGUGAAGAGAAAAGCGACGA--UUUCUCUUGCAGAAUGUUUCAGGUUGCACAUAGCUAUGGUUGCAAGUGCUAGCGCUUUUAAAA---UACAU--GAAUUGU--GUUCUUGAAAUUGAA .(((.((((((((........--)))))))))))....((((((((..(((((....((((.(((.(((((....))))).)))..---..)))--)...)))--)).)))))))).... ( -28.80) >DroYak_CAF1 33123 115 + 1 AUGCAAAAAGAGAAA-UACGA--UUUCUCUUGCAGAAUGUUGCAGGUUGCAAAUAGCAUCGGUUGCAAGAGCUAGCAAUUUUAAAAUAAUACAU--GAAUUGUGUGUUCUUGAAAUUGCA .(((((.((((((((-(...)--))))))))((((....))))...)))))..((((.((........))))))((((((((((...(((((((--.....))))))).)))))))))). ( -33.70) >consensus AUGCAAAGAGAA__A_UACGU__CUUCUCUUGCAGAAUGUUUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCUAGCCAUUCGAAAA___UGCAA__GAGUUGU__GUUCUUGAAAUUGAA ..((.(((((((............)))))))((((..((...))..)))).....)).((((((.(((((((..((.((((...............)))).))..))))))).)))))). (-16.76 = -17.28 + 0.52)



| Location | 16,754,292 – 16,754,399 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -14.28 |
| Energy contribution | -16.92 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16754292 107 + 22224390 UUCAGGUUGCAAAUAGCUUUGGUUGCAAGAGCUAGCCAUUCGAAAA---UGCAC--GAGUUGU--GUUCUUGAAAUUGAA------AUUGCAAUAGCAGAAAUGUAUAUAUACAUAUACA (((..(((((((.....((..(((.(((((((..((.(((((....---....)--)))).))--))))))).)))..))------.)))))))....))).(((((((....))))))) ( -26.00) >DroSec_CAF1 41332 105 + 1 UUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCCAGCCAUUCGAAAA---UGCAAUGCAGUUGU--GUUCUUGAAAUUGAA------AUUGCAAUAGCAGAAAUGUAUAUAUG----UACA .....(((((((.....(((((((.((((((((((((((((.....---.).))))..)))).--))))))).)))))))------.))))))).(((....))).......----.... ( -25.90) >DroSim_CAF1 38495 105 + 1 UUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCCAGCCAUUCGAAAA---UGCAAUGCAGUUGU--GUUCUUGAAAUUGAA------AUUGCAAUAGCAGAAAUGUAUAUAUG----UACA .....(((((((.....(((((((.((((((((((((((((.....---.).))))..)))).--))))))).)))))))------.))))))).(((....))).......----.... ( -25.90) >DroEre_CAF1 32845 98 + 1 UUCAGGUUGCACAUAGCUAUGGUUGCAAGUGCUAGCGCUUUUAAAA---UACAU--GAAUUGU--GUUCUUGAAAUUGAA------AUUGCAAUAGCAGAAUUGUUUAUGU--------- ((((((..(((((....((((.(((.(((((....))))).)))..---..)))--)...)))--)).))))))......------.((((....))))............--------- ( -20.80) >DroYak_CAF1 33160 109 + 1 UGCAGGUUGCAAAUAGCAUCGGUUGCAAGAGCUAGCAAUUUUAAAAUAAUACAU--GAAUUGUGUGUUCUUGAAAUUGCAAUUGCAAUUGCAAUAGCAGAAAUGUAUAUAU--------- (((((.((((((.((((.((........))))))((((((((((...(((((((--.....))))))).))))))))))..)))))))))))...(((....)))......--------- ( -33.20) >consensus UUCAGGUUGCAAAUAGCUUCGGUUGCAAGAGCUAGCCAUUCGAAAA___UGCAA__GAGUUGU__GUUCUUGAAAUUGAA______AUUGCAAUAGCAGAAAUGUAUAUAU_____UACA .....(((((((.....(((((((.(((((((..((.((((...............)))).))..))))))).))))))).......))))))).(((....)))............... (-14.28 = -16.92 + 2.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:17 2006