
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,752,459 – 16,752,574 |
| Length | 115 |
| Max. P | 0.733072 |

| Location | 16,752,459 – 16,752,574 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -19.63 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
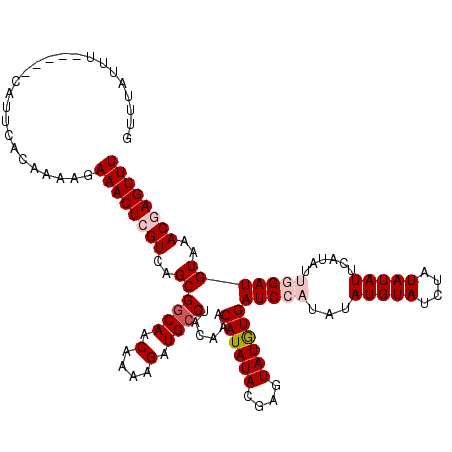
>X_DroMel_CAF1 16752459 115 + 22224390 AAACUCGUUUACAUCCAAUAUGAAUAUAUAGAUACAUAUAUGGAUCAUAUACUCGUAUAUGUUUGUAGGCAUCUUUUGUUGCCGCUGACGAGUUUCUUUUGUGAAUG-----AAAUAAAC (((((((((((((..((((((((.(((((.(((.((....)).)))))))).)))))).))..))))((((........))))...)))))))))............-----........ ( -29.10) >DroSec_CAF1 39525 115 + 1 AAACUCGUUUACAUCAGAUAUGAAUAUAUAGAUACAUAUAUGGAUCACAUACGCGUAUAUGUUUGUAGGCAUCUUUUGUUGCCGCUGACGAGUUUCUUUUGUGAAUG-----AAAUAAAC ....(((((((((..(((.((............((((((((((........).)))))))))((((.((((........))))....)))))).)))..))))))))-----)....... ( -26.30) >DroSim_CAF1 36666 115 + 1 AAACUCGUUUACAUCAGAUAUGAAUAUAUAGAUACAUAUAUGGAUCACAUACGCGUAUAUGUUUGUAGGCAUCUUUUGUUGCCGCUGACGAGUUUCUUUUGUGAAUG-----AAAUAAAC ....(((((((((..(((.((............((((((((((........).)))))))))((((.((((........))))....)))))).)))..))))))))-----)....... ( -26.30) >DroEre_CAF1 31040 107 + 1 AAACUUGUUUACAUCCAAUCUGAAUAUAUAGAUACAU--AUGGAUCACAU------AUAUGUUUGUAGGCAUCUUUUGUUGCCGCUGACGAGUUUCUUUUGUGAAUG-----AAAUAAAC (((((((((..(..............((((((((.((--(((.....)))------)).))))))))((((........)))))..)))))))))............-----........ ( -23.80) >DroYak_CAF1 31219 116 + 1 AAACUUGUUUACAUCCAAUAUGAAUAUAUAGAUACAU--AUGGAUCACAUACU--UAUAUGUUUGUAGGCACCUUUUGUUGGCGCUGACGAGUUUCUUUUGUGAAUGAAAUGAAAUAAAC (((((((((.........((..(((((((((.....(--(((.....)))).)--))))))))..))(((.((.......)).))))))))))))......................... ( -21.90) >consensus AAACUCGUUUACAUCCAAUAUGAAUAUAUAGAUACAUAUAUGGAUCACAUACGCGUAUAUGUUUGUAGGCAUCUUUUGUUGCCGCUGACGAGUUUCUUUUGUGAAUG_____AAAUAAAC (((((((((..(......((((((((((((........((((.....))))....))))))))))))((((.(....).)))))..)))))))))......................... (-19.63 = -20.08 + 0.45)


| Location | 16,752,459 – 16,752,574 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -16.85 |
| Energy contribution | -18.14 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16752459 115 - 22224390 GUUUAUUU-----CAUUCACAAAAGAAACUCGUCAGCGGCAACAAAAGAUGCCUACAAACAUAUACGAGUAUAUGAUCCAUAUAUGUAUCUAUAUAUUCAUAUUGGAUGUAAACGAGUUU ........-----............((((((((....((((.(....).))))((((..((.(((.(((((((((((.((....)).))).)))))))).)))))..)))).)))))))) ( -26.50) >DroSec_CAF1 39525 115 - 1 GUUUAUUU-----CAUUCACAAAAGAAACUCGUCAGCGGCAACAAAAGAUGCCUACAAACAUAUACGCGUAUGUGAUCCAUAUAUGUAUCUAUAUAUUCAUAUCUGAUGUAAACGAGUUU ........-----............((((((((..((((((.(....).)))).......(((((.(((((((((...)))))))))...))))).............))..)))))))) ( -23.10) >DroSim_CAF1 36666 115 - 1 GUUUAUUU-----CAUUCACAAAAGAAACUCGUCAGCGGCAACAAAAGAUGCCUACAAACAUAUACGCGUAUGUGAUCCAUAUAUGUAUCUAUAUAUUCAUAUCUGAUGUAAACGAGUUU ........-----............((((((((..((((((.(....).)))).......(((((.(((((((((...)))))))))...))))).............))..)))))))) ( -23.10) >DroEre_CAF1 31040 107 - 1 GUUUAUUU-----CAUUCACAAAAGAAACUCGUCAGCGGCAACAAAAGAUGCCUACAAACAUAU------AUGUGAUCCAU--AUGUAUCUAUAUAUUCAGAUUGGAUGUAAACAAGUUU ((((((((-----((.((...................((((.(....).)))).......((((------(((((...)))--))))))...........)).)))).))))))...... ( -18.30) >DroYak_CAF1 31219 116 - 1 GUUUAUUUCAUUUCAUUCACAAAAGAAACUCGUCAGCGCCAACAAAAGGUGCCUACAAACAUAUA--AGUAUGUGAUCCAU--AUGUAUCUAUAUAUUCAUAUUGGAUGUAAACAAGUUU ((((((..........(((((..........((..(((((.......)))))..))..((.....--.)).)))))(((((--(((............)))).)))).))))))...... ( -21.10) >consensus GUUUAUUU_____CAUUCACAAAAGAAACUCGUCAGCGGCAACAAAAGAUGCCUACAAACAUAUACGAGUAUGUGAUCCAUAUAUGUAUCUAUAUAUUCAUAUUGGAUGUAAACGAGUUU .........................((((((((..((((((.(....).))))......((((((....))))))(((((...(((((....)))))......)))))))..)))))))) (-16.85 = -18.14 + 1.29)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:08 2006