
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,751,334 – 16,751,481 |
| Length | 147 |
| Max. P | 0.876942 |

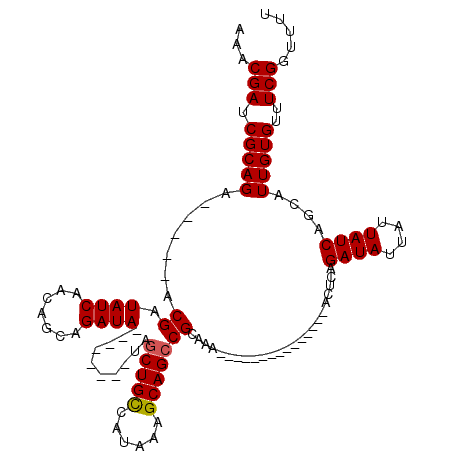
| Location | 16,751,334 – 16,751,441 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16751334 107 - 22224390 AAACGAUCGCAGA-----ACGAUAUCAACAGCAGAUA--------UAGCUGCCAUAAAGCAGCCGCAAAAAAAAAAAGAAUAAAAUUCAGAUAUUAUUAUCAGCAUUGUGUUUCGGUUUU ...(((.(((((.-----...(((((.......))))--------).(((((......))))).((...........((((...)))).((((....)))).)).)))))..)))..... ( -21.50) >DroSec_CAF1 38518 101 - 1 AAACGAUCGCAGA-----ACGAUAUCAACAGCAGAUA--------UAGCUGCCAUAAAGCAGACGCAAAAAAAAAA------GAACUCAGAUAUUAUUAUCAUCAUUGUGUUUCGGUUUU ....((((((((.-----...(((((.......))))--------)..))))......(.((((((((........------(....).((((....))))....))))))))))))).. ( -18.30) >DroEre_CAF1 30061 92 - 1 AAACGAUCGCAGC-----ACGAUAUCAACAGCAGAUA--------UAGCUGUCAUAAAGCAGCCGCAAA---------------AGUCAGAUAUUAUUAUCAGCAUUGUGUUUCGGUUUU ....(((((.(((-----(((((....(((((.....--------..)))))......((....((...---------------.))..((((....)))).)))))))))).))))).. ( -20.90) >DroYak_CAF1 30078 105 - 1 AAACGAUCGCAGCACGGCACGAUAUCAACAGCAGAUAUACAUACAUAGCUGCCAUAAAGCAGCCGCAAA---------------ACUCAGAUAUUAUUAUCAGCAUUGUGUUUCGGUUUU ....(((((.((((((((...(((((.......))))).........(((((......))))).))...---------------.....((((....)))).....)))))).))))).. ( -24.50) >consensus AAACGAUCGCAGA_____ACGAUAUCAACAGCAGAUA________UAGCUGCCAUAAAGCAGCCGCAAA_______________ACUCAGAUAUUAUUAUCAGCAUUGUGUUUCGGUUUU ...(((.(((((.......((.((((.......))))..........(((((......)))))))........................((((....))))....)))))..)))..... (-15.64 = -15.70 + 0.06)

| Location | 16,751,374 – 16,751,481 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -23.75 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16751374 107 + 22224390 UUCUUUUUUUUUUUGCGGCUGCUUUAUGGCAGCUA--------UAUCUGCUGUUGAUAUCGU-----UCUGCGAUCGUUUGCACAACAACUUCGUCUUGAAUGACAGCGUAGAAGACGGC ........(((((((((((((((....))))))).--------.....((((((......((-----(.(((((....))))).)))...(((.....))).)))))))))))))).... ( -31.40) >DroSec_CAF1 38556 103 + 1 ----UUUUUUUUUUGCGUCUGCUUUAUGGCAGCUA--------UAUCUGCUGUUGAUAUCGU-----UCUGCGAUCGUUUGCACAACAACUUCGUCUUGAAUGACAGCGUUGAAGACGGC ----............(.(((((....)))))).(--------((((.......))))).((-----(.(((((....))))).)))....(((((((.((((....)))).))))))). ( -26.40) >DroEre_CAF1 30097 96 + 1 -----------UUUGCGGCUGCUUUAUGACAGCUA--------UAUCUGCUGUUGAUAUCGU-----GCUGCGAUCGUUUGCACAAUAACUUCGUCUUGAAUGACAGCGCAGAAGACGGC -----------......((((((((..((((((..--------.....))))))......((-----(((((((....))))...........(((......)))))))).)))).)))) ( -28.10) >DroYak_CAF1 30114 109 + 1 -----------UUUGCGGCUGCUUUAUGGCAGCUAUGUAUGUAUAUCUGCUGUUGAUAUCGUGCCGUGCUGCGAUCGUUUGCACAGCAACUUCGUCUUGAAUGACAGCGUAGAAGACGGC -----------.(((((((((((....)))))))..(((((.(((((.......)))))))))).((((.((....))..)))).))))..(((((((..(((....)))..))))))). ( -36.80) >consensus ___________UUUGCGGCUGCUUUAUGGCAGCUA________UAUCUGCUGUUGAUAUCGU_____GCUGCGAUCGUUUGCACAACAACUUCGUCUUGAAUGACAGCGUAGAAGACGGC ................(((((((....)))))))..............((((((.....(((.......(((((....)))))..........(((......))).))).....)))))) (-23.75 = -24.25 + 0.50)



| Location | 16,751,374 – 16,751,481 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16751374 107 - 22224390 GCCGUCUUCUACGCUGUCAUUCAAGACGAAGUUGUUGUGCAAACGAUCGCAGA-----ACGAUAUCAACAGCAGAUA--------UAGCUGCCAUAAAGCAGCCGCAAAAAAAAAAAGAA ((..........((((((......)))((.((((((.(((........))).)-----))))).))...))).....--------..(((((......))))).)).............. ( -26.70) >DroSec_CAF1 38556 103 - 1 GCCGUCUUCAACGCUGUCAUUCAAGACGAAGUUGUUGUGCAAACGAUCGCAGA-----ACGAUAUCAACAGCAGAUA--------UAGCUGCCAUAAAGCAGACGCAAAAAAAAAA---- ((.((((.....((((((......)))((.((((((.(((........))).)-----))))).))...))).....--------..(((.......)))))))))..........---- ( -26.10) >DroEre_CAF1 30097 96 - 1 GCCGUCUUCUGCGCUGUCAUUCAAGACGAAGUUAUUGUGCAAACGAUCGCAGC-----ACGAUAUCAACAGCAGAUA--------UAGCUGUCAUAAAGCAGCCGCAAA----------- ((.....(((((..((((......))))..(.((((((((...........))-----)))))).)....)))))..--------..(((((......))))).))...----------- ( -26.20) >DroYak_CAF1 30114 109 - 1 GCCGUCUUCUACGCUGUCAUUCAAGACGAAGUUGCUGUGCAAACGAUCGCAGCACGGCACGAUAUCAACAGCAGAUAUACAUACAUAGCUGCCAUAAAGCAGCCGCAAA----------- (((((.....((..((((......))))..)).((((((........))))))))))).(((((((.......))))).........(((((......)))))))....----------- ( -29.40) >consensus GCCGUCUUCUACGCUGUCAUUCAAGACGAAGUUGUUGUGCAAACGAUCGCAGA_____ACGAUAUCAACAGCAGAUA________UAGCUGCCAUAAAGCAGCCGCAAA___________ ((((((((..............))))))...(((((((.......((((..........))))....))))))).............(((((......))))).)).............. (-18.92 = -19.04 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:06 2006