
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,749,414 – 16,749,586 |
| Length | 172 |
| Max. P | 0.999952 |

| Location | 16,749,414 – 16,749,534 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16749414 120 + 22224390 GGUCGCCAAAAGAUUUUAAAUCAUUAUUCACUCGAAUUCCCUAACGUACCCGCAAUUUUGAUGUCGGAUAAUUUGGCACAAAUGGGGUUUUGUGGGUGGUCCACACCCACACGCCCAUCU .((.((((((.(((.....))).(((((((((((((....................))))).)).)))))))))))))).....((((..((((((((.....)))))))).)))).... ( -35.25) >DroSec_CAF1 36630 120 + 1 GGUCGCCAAAAGAUUUUAAAUCAUUAUUCACUCGAAUUCCCUAACGUACCCGUAAUUUUUAUGUCGGAUAAUUUGGCACAAAUGGGGUUUUGUGGGUGGUCCACACCCACACGCCCAUCU .((.((((((.(((.....))).((((((((..(((.......(((....)))....)))..)).)))))))))))))).....((((..((((((((.....)))))))).)))).... ( -33.30) >DroSim_CAF1 34982 120 + 1 GGUCGCCAAAAGAUUUUAAAUCAUUAUUCACUCGAAUUCCCUAACGUACCCGCAAUUUUUAUGUCGGAUAAUUUGGCGCAAAUGGGGUUUUGUGGGUGGUCCACACCCACACGCCCAUCU .(.(((((((.(((.....)))...........................(((((.......)).)))....)))))))).....((((..((((((((.....)))))))).)))).... ( -34.80) >DroEre_CAF1 28278 120 + 1 GGUCGCUUACAGAUUUUAAAUCAUUAUUCACUUGAGUUCCCUUACGUACCUGCAAUUUUUAUGUCGGAUAAUUUGGCACAAAUGGCAUUUUGUGGGUGGUCCACACCCACACACCCAUCU (((((......(((.....)))...((((....)))).......)).)))(((....(((.(((((((...))))))).)))..)))...((((((((.....))))))))......... ( -25.80) >DroYak_CAF1 28144 120 + 1 GGUGGCUUGUAGAUUUGAAAUCAUUAUUCACUUGACUUUCCUUACGUACCCGCAAUUUUGAUGUCGGCUAAUUUGGCACAAAUGGCAUUUUGUGGGUGGUCCACACCCACACGCCCAUCU .((((..(((((....(((((((.........))).)))).)))))...))))......((((..(((.....((.((....)).))...((((((((.....)))))))).))))))). ( -31.80) >consensus GGUCGCCAAAAGAUUUUAAAUCAUUAUUCACUCGAAUUCCCUAACGUACCCGCAAUUUUUAUGUCGGAUAAUUUGGCACAAAUGGGGUUUUGUGGGUGGUCCACACCCACACGCCCAUCU .((.(((((..(((.....)))...........................(((((.......)).))).....)))))))..(((((....((((((((.....))))))))..))))).. (-26.76 = -26.68 + -0.08)

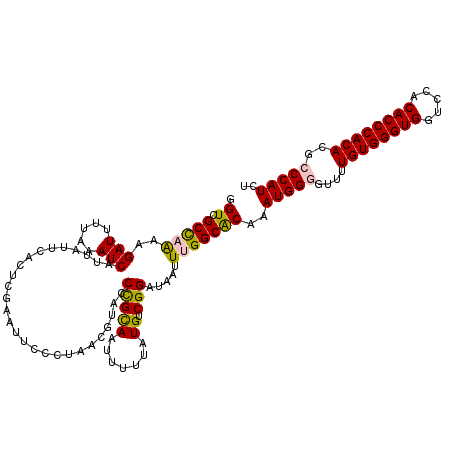

| Location | 16,749,414 – 16,749,534 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -26.94 |
| Energy contribution | -29.50 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16749414 120 - 22224390 AGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUUUGUGCCAAAUUAUCCGACAUCAAAAUUGCGGGUACGUUAGGGAAUUCGAGUGAAUAAUGAUUUAAAAUCUUUUGGCGACC (((((((..((((((((.....))))))))....))))))).(((((((.((((((.((.......)))))))).............((.(((((....)))))...)).)))))))... ( -38.50) >DroSec_CAF1 36630 120 - 1 AGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUUUGUGCCAAAUUAUCCGACAUAAAAAUUACGGGUACGUUAGGGAAUUCGAGUGAAUAAUGAUUUAAAAUCUUUUGGCGACC (((((((..((((((((.....))))))))....))))))).(((((((.((((((............)))))).............((.(((((....)))))...)).)))))))... ( -36.00) >DroSim_CAF1 34982 120 - 1 AGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUUUGCGCCAAAUUAUCCGACAUAAAAAUUGCGGGUACGUUAGGGAAUUCGAGUGAAUAAUGAUUUAAAAUCUUUUGGCGACC (((((((..((((((((.....))))))))....))))))).(((((((.((((((.((.......)))))))).............((.(((((....)))))...)).)))))))... ( -40.30) >DroEre_CAF1 28278 120 - 1 AGAUGGGUGUGUGGGUGUGGACCACCCACAAAAUGCCAUUUGUGCCAAAUUAUCCGACAUAAAAAUUGCAGGUACGUAAGGGAACUCAAGUGAAUAAUGAUUUAAAAUCUGUAAGCGACC ((((((...((((((((.....)))))))).....)))))).(((....((((.....))))...((((((((.(....)..........(((((....)))))..)))))))))))... ( -26.70) >DroYak_CAF1 28144 120 - 1 AGAUGGGCGUGUGGGUGUGGACCACCCACAAAAUGCCAUUUGUGCCAAAUUAGCCGACAUCAAAAUUGCGGGUACGUAAGGAAAGUCAAGUGAAUAAUGAUUUCAAAUCUACAAGCCACC .((((((((((((((((.....))))))))...))))..(((.((.......)))))))))....(((((....)))))((...((....((((.......)))).....))...))... ( -32.00) >consensus AGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUUUGUGCCAAAUUAUCCGACAUAAAAAUUGCGGGUACGUUAGGGAAUUCGAGUGAAUAAUGAUUUAAAAUCUUUUGGCGACC (((((((..((((((((.....))))))))....))))))).((((((..((((((.((.......))))))))(((((....((....))...)))))............))))))... (-26.94 = -29.50 + 2.56)

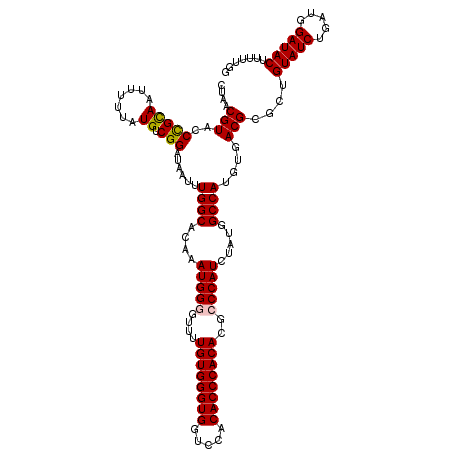
| Location | 16,749,454 – 16,749,574 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -32.98 |
| Energy contribution | -33.06 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16749454 120 + 22224390 CUAACGUACCCGCAAUUUUGAUGUCGGAUAAUUUGGCACAAAUGGGGUUUUGUGGGUGGUCCACACCCACACGCCCAUCUAUGGCCAUGUGACGUGCUGUAUCUGAUGGAUACUUUUUGG .....((((.((((..((((....)))).....((((.......((((..((((((((.....)))))))).)))).......))))))))..)))).(((((.....)))))....... ( -39.34) >DroSec_CAF1 36670 120 + 1 CUAACGUACCCGUAAUUUUUAUGUCGGAUAAUUUGGCACAAAUGGGGUUUUGUGGGUGGUCCACACCCACACGCCCAUCUAUGGCCAUGUGACGCGCUGUAUCUGAUGGAUACUUUUUGG .....(((.(((((.....)))((((((((...((((.......((((..((((((((.....)))))))).)))).......)))).(((...)))..)))))))))).)))....... ( -37.94) >DroSim_CAF1 35022 120 + 1 CUAACGUACCCGCAAUUUUUAUGUCGGAUAAUUUGGCGCAAAUGGGGUUUUGUGGGUGGUCCACACCCACACGCCCAUCUAUGGCCAUGUGACGCGCUGUAUCUGAUGGAUACUUUUUGG .....(((.((...........((((((((....(((((.....((((..((((((((.....)))))))).)))).(((((....))).)).))))).)))))))))).)))....... ( -38.80) >DroEre_CAF1 28318 120 + 1 CUUACGUACCUGCAAUUUUUAUGUCGGAUAAUUUGGCACAAAUGGCAUUUUGUGGGUGGUCCACACCCACACACCCAUCUAAGGCCAUGUGACGCGCUGUAUCUGAUGGAUACUUUUUGG .((((((.((.(((.......))).)).......(((....((((.....((((((((.....))))))))...)))).....)))))))))......(((((.....)))))....... ( -33.00) >DroYak_CAF1 28184 120 + 1 CUUACGUACCCGCAAUUUUGAUGUCGGCUAAUUUGGCACAAAUGGCAUUUUGUGGGUGGUCCACACCCACACGCCCAUCUAUGGCCAUGUGACGCGCUGUAUCUGAUGGAUACUUUUUGG .....(((.(((((.....((((.((((..((.((((.((.((((.....((((((((.....))))))))...))))...)))))).)).....)))))))))).))).)))....... ( -34.30) >consensus CUAACGUACCCGCAAUUUUUAUGUCGGAUAAUUUGGCACAAAUGGGGUUUUGUGGGUGGUCCACACCCACACGCCCAUCUAUGGCCAUGUGACGCGCUGUAUCUGAUGGAUACUUUUUGG ....(((..(((((.......)).)))......((((....(((((....((((((((.....))))))))..))))).....))))....)))....(((((.....)))))....... (-32.98 = -33.06 + 0.08)


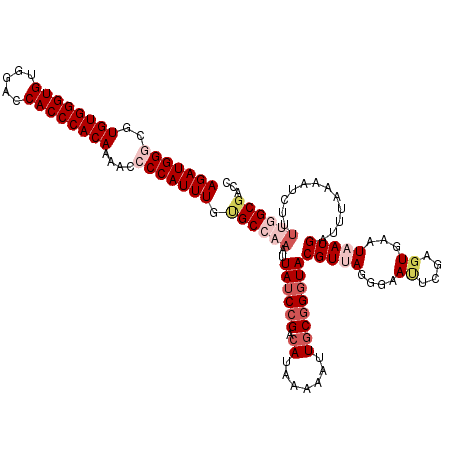
| Location | 16,749,454 – 16,749,574 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -37.98 |
| Energy contribution | -39.02 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.81 |
| SVM RNA-class probability | 0.999952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16749454 120 - 22224390 CCAAAAAGUAUCCAUCAGAUACAGCACGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUUUGUGCCAAAUUAUCCGACAUCAAAAUUGCGGGUACGUUAG .......(((((.....)))))...(((.....((((.(((((((((..((((((((.....))))))))....)))))))))))))...((((((.((.......)))))))))))... ( -44.20) >DroSec_CAF1 36670 120 - 1 CCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUUUGUGCCAAAUUAUCCGACAUAAAAAUUACGGGUACGUUAG .......(((((.....)))))((((.......((((.(((((((((..((((((((.....))))))))....)))))))))))))...((((((............)))))))))).. ( -43.00) >DroSim_CAF1 35022 120 - 1 CCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUUUGCGCCAAAUUAUCCGACAUAAAAAUUGCGGGUACGUUAG .......(((((.....)))))((((.......((((..((((((((..((((((((.....))))))))....)))))))).))))...((((((.((.......)))))))))))).. ( -42.80) >DroEre_CAF1 28318 120 - 1 CCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCUUAGAUGGGUGUGUGGGUGUGGACCACCCACAAAAUGCCAUUUGUGCCAAAUUAUCCGACAUAAAAAUUGCAGGUACGUAAG .......(((((.....)))))...((((....((((..(((((((...((((((((.....)))))))).....))))))).))))......((..((.......))..)).))))... ( -35.30) >DroYak_CAF1 28184 120 - 1 CCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAAUGCCAUUUGUGCCAAAUUAGCCGACAUCAAAAUUGCGGGUACGUAAG .......(((((.....)))))...((((.((.((((.((((((((...((((((((.....)))))))).....))))))))))))......(((.((.......)))))))))))... ( -41.10) >consensus CCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUUUGUGCCAAAUUAUCCGACAUAAAAAUUGCGGGUACGUUAG .......(((((.....)))))...((((.((.((((.(((((((((..((((((((.....))))))))....)))))))))))))......(((.((.......)))))))))))... (-37.98 = -39.02 + 1.04)



| Location | 16,749,494 – 16,749,586 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.02 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16749494 92 - 22224390 ----------------------------CAAUUCCAAAUCCCAAAAAGUAUCCAUCAGAUACAGCACGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUU ----------------------------...................(((((.....))))).....(((....)))....((((((..((((((((.....))))))))....)))))) ( -26.60) >DroSec_CAF1 36710 113 - 1 GUUCCCCUGUUCCGCAAGC-------CUCAAUCCCAAAUCCCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUU ....((.(((..(((..(.-------..)..................(((((.....)))))...)))..))).)).....((((((..((((((((.....))))))))....)))))) ( -31.00) >DroSim_CAF1 35062 113 - 1 UUUCCCCUGUUCCGCAAAC-------CUCAAUCCCAAAUCCCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUU ....((.(((..(((....-------....((.....))........(((((.....)))))...)))..))).)).....((((((..((((((((.....))))))))....)))))) ( -30.40) >DroEre_CAF1 28358 112 - 1 GU-CGCCUGUUCCCCAAUC-------CGCAAUCCCAAAUCCCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCUUAGAUGGGUGUGUGGGUGUGGACCACCCACAAAAUGCCAUU ..-.((..........(((-------((((..((((...........(((((.....))))).....(((....)))......))))..)))))))((((.....)))).....)).... ( -27.20) >DroYak_CAF1 28224 119 - 1 GU-UUCCCAAUCCCCAAUCCACAAACCGCAAUCCCAAAUCCCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAAUGCCAUU ..-........................(((.........(((.....(((((.....))))).(((((((.(((........)))))))))))))(((((.....)))))...))).... ( -28.50) >consensus GU_CCCCUGUUCCCCAAUC_______CGCAAUCCCAAAUCCCAAAAAGUAUCCAUCAGAUACAGCGCGUCACAUGGCCAUAGAUGGGCGUGUGGGUGUGGACCACCCACAAAACCCCAUU .......................................(((.....(((((.....))))).(((((((.(((........)))))))))))))(((((.....))))).......... (-25.34 = -25.02 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:02 2006