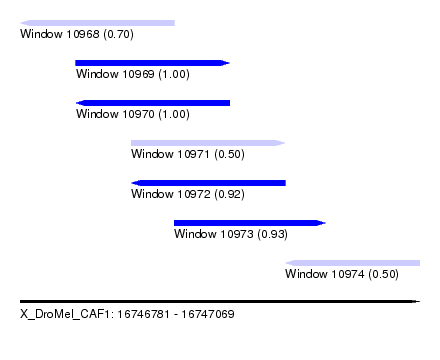
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,746,781 – 16,747,069 |
| Length | 288 |
| Max. P | 0.999975 |

| Location | 16,746,781 – 16,746,892 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -20.44 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746781 111 - 22224390 ACAACAAAAAAA--------C-GGGGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAUUACGUUUUAACAAAUCAAAUAUAUUUUAAGUGCAAAAUAA ........((((--------(-(.(((((((((......)))))))))(((((((....(((....))).))))))).....)))))).............((((((......)))))). ( -21.80) >DroSec_CAF1 34105 111 - 1 ACAACAAAAAAA--------C-GGUGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCUUUUGGCAGUUUGCCUAUUACGUUUUAACAAAUCAAAUAUAUUUUAAGUGCAAAAUAA ........((((--------(-(..((((((((......)))))))).(((((((....(((....))).))))))).....)))))).............((((((......)))))). ( -20.70) >DroSim_CAF1 32426 111 - 1 ACAACAAAAAAA--------C-GGUGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAUUACGUUUUAACAAAUCAAAUAUAUUUUAAGUGCAAAAUAA ........((((--------(-(..((((((((......)))))))).(((((((....(((....))).))))))).....)))))).............((((((......)))))). ( -20.50) >DroEre_CAF1 25771 110 - 1 ACAAGAAAAAAA----------GAGGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAUUACGUUUUAACAAAUCAAAUAUAUUUUAAGUGCAAAAUAA ............----------(((((((((((......)))))))))(((((((....(((....))).))))))).................)).....((((((......)))))). ( -17.00) >DroYak_CAF1 25559 120 - 1 ACAAAAAAAAAAAAAAGAAACGGGGGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAUUACGUUUUAACAAAUCAAAUAUAUUUUAAGUGCAAAAUAA ................((((((..(((((((((......)))))))))(((((((....(((....))).))))))).....)))))).............((((((......)))))). ( -22.20) >consensus ACAACAAAAAAA________C_GGGGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAUUACGUUUUAACAAAUCAAAUAUAUUUUAAGUGCAAAAUAA ........................(((((((((......)))))))))(((((((....(((....))).)))))))........................((((((......)))))). (-15.78 = -16.18 + 0.40)



| Location | 16,746,821 – 16,746,932 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -24.93 |
| Energy contribution | -25.29 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746821 111 + 22224390 AUAGGCAAACUGCCAAGUGGCUAAUGUUUACCAAAACAAUUUGAUUCAAUUAUUCCCC-G--------UUUUUUUGUUGUAUUUUUGUGUCUGGGCAAAUAGCCAACUGGCAUUUUCCGC ...((.(((.(((((..((((((.(((...(((..((((...(((.((((........-.--------.......)))).))).))))...))))))..))))))..))))).))))).. ( -27.39) >DroSec_CAF1 34145 111 + 1 AUAGGCAAACUGCCAAAAGGCUAAUGUUUACCAAAACAAUUUGAUUCAAUUAUUCACC-G--------UUUUUUUGUUGUAUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGC ...((.(((.(((((...((((...((((.(((..((((...(((.((((........-.--------.......)))).))).))))...)))..))))))))...))))).))))).. ( -24.59) >DroSim_CAF1 32466 111 + 1 AUAGGCAAACUGCCAAGUGGCUAAUGUUUACCAAAACAAUUUGAUUCAAUUAUUCACC-G--------UUUUUUUGUUGUAUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGC ...((.(((.(((((..(((((...((((.(((..((((...(((.((((........-.--------.......)))).))).))))...)))..)))))))))..))))).))))).. ( -27.99) >DroEre_CAF1 25811 110 + 1 AUAGGCAAACUGCCAAGUGGCUAAUGUUUACCAAAACAAUUUGAUUCAAUUAUUCCUC----------UUUUUUUCUUGUAUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGC ...((.(((.(((((..(((((...((((.(((..((((...(((.(((.........----------........))).))).))))...)))..)))))))))..))))).))))).. ( -26.03) >DroYak_CAF1 25599 120 + 1 AUAGGCAAACUGCCAAGUGGCUAAUGUUUACCAAAACAAUUUGAUUCAAUUAUUCCCCCGUUUCUUUUUUUUUUUUUUGUAUUUUUGUGUCUGGGCAAAUAGCCAACUGGCAUUUUCCGC ...((.(((.(((((..((((((.(((...(((..((((...(((.(((...........................))).))).))))...))))))..))))))..))))).))))).. ( -25.73) >consensus AUAGGCAAACUGCCAAGUGGCUAAUGUUUACCAAAACAAUUUGAUUCAAUUAUUCCCC_G________UUUUUUUGUUGUAUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGC ...((.(((.(((((..(((((...((((.(((..((((...(((.((((.........................)))).))).))))...)))..)))))))))..))))).))))).. (-24.93 = -25.29 + 0.36)



| Location | 16,746,821 – 16,746,932 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -28.50 |
| Energy contribution | -28.46 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.14 |
| SVM RNA-class probability | 0.999975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746821 111 - 22224390 GCGGAAAAUGCCAGUUGGCUAUUUGCCCAGACACAAAAAUACAACAAAAAAA--------C-GGGGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAU (((((...((((((.((((((((((((.((((.........(..(.......--------.-)..)...((((......)))))))).))))))...)))))).)))))).))))).... ( -30.40) >DroSec_CAF1 34145 111 - 1 GCGGAAAAUGCCAGUUGGCUGUUUGCCCAGACACAAAAAUACAACAAAAAAA--------C-GGUGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCUUUUGGCAGUUUGCCUAU (((((...((((((..(((((((((((....(((..................--------.-.)))(((((((......)))))))..)))))))...))))..)))))).))))).... ( -31.25) >DroSim_CAF1 32466 111 - 1 GCGGAAAAUGCCAGUUGGCUGUUUGCCCAGACACAAAAAUACAACAAAAAAA--------C-GGUGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAU (((((...((((((.((((((((((((....(((..................--------.-.)))(((((((......)))))))..)))))))...))))).)))))).))))).... ( -31.75) >DroEre_CAF1 25811 110 - 1 GCGGAAAAUGCCAGUUGGCUGUUUGCCCAGACACAAAAAUACAAGAAAAAAA----------GAGGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAU (((((...((((((.((((((((((((.(((((........(((........----------.........)))........))))).)))))))...))))).)))))).))))).... ( -31.72) >DroYak_CAF1 25599 120 - 1 GCGGAAAAUGCCAGUUGGCUAUUUGCCCAGACACAAAAAUACAAAAAAAAAAAAAAGAAACGGGGGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAU (((((...((((((.(((((((((.(((.................................))).))))).....((((((....))))))........)))).)))))).))))).... ( -30.01) >consensus GCGGAAAAUGCCAGUUGGCUGUUUGCCCAGACACAAAAAUACAACAAAAAAA________C_GGGGAAUAAUUGAAUCAAAUUGUUUUGGUAAACAUUAGCCACUUGGCAGUUUGCCUAU (((((...((((((.((((((((((((.(((((........(((...........................)))........))))).))))))...)))))).)))))).))))).... (-28.50 = -28.46 + -0.04)



| Location | 16,746,861 – 16,746,972 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.16 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746861 111 + 22224390 UUGAUUCAAUUAUUCCCC-G--------UUUUUUUGUUGUAUUUUUGUGUCUGGGCAAAUAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUCGG ................((-(--------..(((((((((...(((((((((.(.(((....((((...(((.......)))...))))....))).)))))))))).))))))))).))) ( -29.60) >DroSec_CAF1 34185 111 + 1 UUGAUUCAAUUAUUCACC-G--------UUUUUUUGUUGUAUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUCGG ................((-(--------..(((((((((...(((((((((.(.(((....((((...(((.......)))...))))....))).)))))))))).))))))))).))) ( -29.80) >DroSim_CAF1 32506 111 + 1 UUGAUUCAAUUAUUCACC-G--------UUUUUUUGUUGUAUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUCGG ................((-(--------..(((((((((...(((((((((.(.(((....((((...(((.......)))...))))....))).)))))))))).))))))))).))) ( -29.80) >DroEre_CAF1 25851 110 + 1 UUGAUUCAAUUAUUCCUC----------UUUUUUUCUUGUAUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUUGG ..................----------............(((((((((((..(((.....)))....))).............((((((..(((.....)))))))))))))))))... ( -24.00) >DroYak_CAF1 25639 120 + 1 UUGAUUCAAUUAUUCCCCCGUUUCUUUUUUUUUUUUUUGUAUUUUUGUGUCUGGGCAAAUAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAGAAAUCGG .................((((((((.............((......(((((..(((.....)))....))))).....))....((((((..(((.....)))))))))..))))).))) ( -27.00) >consensus UUGAUUCAAUUAUUCCCC_G________UUUUUUUGUUGUAUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUCGG .....................................((.(((((((((((..(((.....)))....))).............((((((..(((.....))))))))))))))))))). (-24.48 = -24.16 + -0.32)



| Location | 16,746,861 – 16,746,972 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746861 111 - 22224390 CCGAUUUUUGUUGGCCCUGCAUCGCACAGUGGGCCAGAAAGCGGAAAAUGCCAGUUGGCUAUUUGCCCAGACACAAAAAUACAACAAAAAAA--------C-GGGGAAUAAUUGAAUCAA (((.(((((((((((((.((........))))))).......((.(((((((....))).))))..))..............))))))))..--------)-))................ ( -27.50) >DroSec_CAF1 34185 111 - 1 CCGAUUUUUGUUGGCCCUGCAUCGCACAGUGGGCCAGAAAGCGGAAAAUGCCAGUUGGCUGUUUGCCCAGACACAAAAAUACAACAAAAAAA--------C-GGUGAAUAAUUGAAUCAA (((.(((((((((((((.((........))))))).....(((((....(((....)))..)))))................))))))))..--------)-))................ ( -27.40) >DroSim_CAF1 32506 111 - 1 CCGAUUUUUGUUGGCCCUGCAUCGCACAGUGGGCCAGAAAGCGGAAAAUGCCAGUUGGCUGUUUGCCCAGACACAAAAAUACAACAAAAAAA--------C-GGUGAAUAAUUGAAUCAA (((.(((((((((((((.((........))))))).....(((((....(((....)))..)))))................))))))))..--------)-))................ ( -27.40) >DroEre_CAF1 25851 110 - 1 CCAAUUUUUGUUGGCCCUGCAUCGCACAGUGGGCCAGAAAGCGGAAAAUGCCAGUUGGCUGUUUGCCCAGACACAAAAAUACAAGAAAAAAA----------GAGGAAUAAUUGAAUCAA ...((((((((((((((.((........))))))))....(((((....(((....)))..)))))......))))))))............----------.................. ( -25.30) >DroYak_CAF1 25639 120 - 1 CCGAUUUCUGUUGGCCCUGCAUCGCACAGUGGGCCAGAAAGCGGAAAAUGCCAGUUGGCUAUUUGCCCAGACACAAAAAUACAAAAAAAAAAAAAAGAAACGGGGGAAUAAUUGAAUCAA (((.(((((.(((((((.((........))))))))).....((.(((((((....))).))))..))...........................))))))))................. ( -28.10) >consensus CCGAUUUUUGUUGGCCCUGCAUCGCACAGUGGGCCAGAAAGCGGAAAAUGCCAGUUGGCUGUUUGCCCAGACACAAAAAUACAACAAAAAAA________C_GGGGAAUAAUUGAAUCAA ...((((((((((((((.((........))))))))....(((((....(((....)))..)))))......))))))))........................................ (-24.38 = -24.58 + 0.20)


| Location | 16,746,892 – 16,747,001 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -26.90 |
| Energy contribution | -27.54 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746892 109 + 22224390 AUUUUUGUGUCUGGGCAAAUAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUCGGAAUCGGAAAAAAAGCUCGUGAUCUUUCUC----------- ........((((((((.....(((....))).(((((((.((((((((((..(((.....))))))))).........)))).)))))))...))))).))).......----------- ( -32.90) >DroSec_CAF1 34216 108 + 1 AUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUCGGAAUCGGAAA-AAAGCUCGUGAUCUUUCUC----------- ........((((((((.....(((....))).(((((((.((((((((((..(((.....))))))))).........)))).))))))-)..))))).))).......----------- ( -32.90) >DroSim_CAF1 32537 108 + 1 AUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUCGGAAUCGGAAA-AAAGCUCGUGAUCUUUCUC----------- ........((((((((.....(((....))).(((((((.((((((((((..(((.....))))))))).........)))).))))))-)..))))).))).......----------- ( -32.90) >DroEre_CAF1 25881 113 + 1 AUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUUGG------AAA-AAAGCUCGUGAUCUUUCUCUUCGCUCACUU ......(((...((((.....(((....))).(((((((.(((.((((((..(((.....)))))))))....))).)))------)))-)..))))((((.........)))).))).. ( -32.50) >DroYak_CAF1 25679 113 + 1 AUUUUUGUGUCUGGGCAAAUAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAGAAAUCGG------AAA-AAAGCUCGUGAUCUUUCUCUUCGCUCACUU ......(((...((((.....(((....))).(((((((.((((((((((..(((.....))))))))...))))).)))------)))-)..))))((((.........)))).))).. ( -37.50) >consensus AUUUUUGUGUCUGGGCAAACAGCCAACUGGCAUUUUCCGCUUUCUGGCCCACUGUGCGAUGCAGGGCCAACAAAAAUCGGAAUCGGAAA_AAAGCUCGUGAUCUUUCUC___________ ........((((((((.....(((....)))...(((((.(((.((((((..(((.....)))))))))....))).)))))...........))))).))).................. (-26.90 = -27.54 + 0.64)

| Location | 16,746,972 – 16,747,069 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -32.56 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746972 97 - 22224390 AUGCCGUCGACGUCGCUGCCGGCGCCUCUGUCGGCGUCGCAGCCGGCGGCAGAAGAGAGCGAUGCGAA-----------------------GAGAAAGAUCACGAGCUUUUUUUCCGAUU .((((((((.....(((((.((((((......)))))))))))))))))))(((((((((....((..-----------------------((......)).)).)))))))))...... ( -41.40) >DroSec_CAF1 34296 96 - 1 AUGCCGUCGACGUCGCUGCCGGCGCCGCUGUCGACGUCGCAGCCGGCGGCAGAAGAGAGCGAUGCGAA-----------------------GAGAAAGAUCACGAGCUUU-UUUCCGAUU .((((((((.....(((((.((((.((....)).)))))))))))))))))(((((((((....((..-----------------------((......)).)).)))))-))))..... ( -37.80) >DroSim_CAF1 32617 96 - 1 AUGCCGUCGACGUCGCUGCCGGCGCCGCUGUCGGCGUCGCAGCCGGCGGCAGAAGAGAGCGAUGCGAA-----------------------GAGAAAGAUCACGAGCUUU-UUUCCGAUU .((((((((.....(((((.(((((((....))))))))))))))))))))(((((((((....((..-----------------------((......)).)).)))))-))))..... ( -44.90) >DroEre_CAF1 25961 107 - 1 AUGCCGUCGACGUCGCUGCCGGCGUCGCUGUCGGCGUCGCAGCCGGCGGCAGAAGAGAGCGAUGCGAAGAG------AUGAAGUGAGCGAAGAGAAAGAUCACGAGCUUU-UUU------ .((((((((.....(((((.(((((((....)))))))))))))))))))).((((((((....(.....)------.....((((.(.........).))))..)))))-)))------ ( -40.10) >DroYak_CAF1 25759 113 - 1 AUGCCGUCGACGUCGCUGCCGGCGUCGCUGUUGGCGUCGCAGCCGGCUGCAGAAGAGAGCGAUGCGAAGAGAUGGAGAUGAAGUGAGCGAAGAGAAAGAUCACGAGCUUU-UUU------ ..((((((..((((.((....((((((((.((..(...((((....)))).)..)).))))))))..)).))))..))))..((((.(.........).))))..))...-...------ ( -38.70) >consensus AUGCCGUCGACGUCGCUGCCGGCGCCGCUGUCGGCGUCGCAGCCGGCGGCAGAAGAGAGCGAUGCGAA_______________________GAGAAAGAUCACGAGCUUU_UUUCCGAUU ..(((((((.((((((((((((((....)))))))(((((.....))))).......))))))))))........................((......)).)).))............. (-32.56 = -32.80 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:53 2006