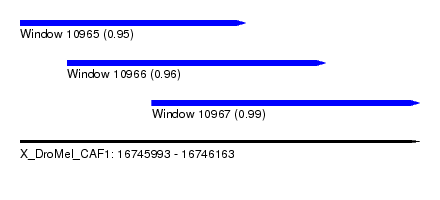
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,745,993 – 16,746,163 |
| Length | 170 |
| Max. P | 0.993720 |

| Location | 16,745,993 – 16,746,089 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 85.30 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16745993 96 + 22224390 ACAAGCACAAGCACACACUCACACUCUUACACACGGAUCGGAUGCUCCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGC ....((....))........................(((.((((((((((....))))....)))))).)))..((((.(((....)))))))... ( -23.50) >DroSec_CAF1 33345 85 + 1 AAAAGCAGA----------CACACUCUCACACAA-AAUCGGAUGCUCCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGC ....((...----------...............-.(((.((((((((((....))))....)))))).)))..((((.(((....))))))).)) ( -23.10) >DroSim_CAF1 31679 85 + 1 ACAAGCACA----------CACACUCUCACACAC-AAUCGGAUGCUCCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGC ....((...----------...............-.(((.((((((((((....))))....)))))).)))..((((.(((....))))))).)) ( -23.10) >DroEre_CAF1 25003 83 + 1 AUAAGCACA------CACUC------ACACACAC-AAUCGGAUACACCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGC ....((.((------(....------........-....(....).((((....))))..))))).((((....((((....)))).))))..... ( -17.30) >consensus ACAAGCACA__________CACACUCUCACACAC_AAUCGGAUGCUCCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGC ...(((..............................(((.((((((((((....))))....)))))).)))..((((....)))))))....... (-19.33 = -19.82 + 0.50)



| Location | 16,746,013 – 16,746,123 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -32.43 |
| Energy contribution | -32.99 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746013 110 + 22224390 ACACUCUUACACACGGAUCGGAUGCUCCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCGCCGCUGGAAGCG------AUUGGCUGGCGUGUGAAUCCC .........(((((..(((.((((((((((....))))....)))))).)))..((((....))))(((....)))(((((..(.....------.)..)).))))))))...... ( -38.60) >DroSec_CAF1 33355 115 + 1 ACACUCUCACACAA-AAUCGGAUGCUCCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCGCCGCUGGAAGCGGUUUGGAUUGGCUGGCGUGUGAAUCCC ......((((((..-.(((.((((((((((....))))....)))))).)))..........(..(((..(.....(((((.....))))).....)..)))..)))))))..... ( -41.70) >DroSim_CAF1 31689 109 + 1 ACACUCUCACACAC-AAUCGGAUGCUCCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCGCCGCUGGAAGCG------AUUGGCUGGCGUGUGAAUCCC ......((((((..-.(((.((((((((((....))))....)))))).)))..((((....))))(((....)))(((((..(.....------.)..)).)))))))))..... ( -39.70) >DroEre_CAF1 25017 103 + 1 ------ACACACAC-AAUCGGAUACACCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCACCGCUGGAAGCG------AUUGGCUGGCGUGUGAAUCCC ------.(((((.(-(((((......((((....))))..(((((((..((.....))..)))...(((....))).))))......))------))))(....))))))...... ( -33.50) >consensus ACACUCUCACACAC_AAUCGGAUGCUCCACGAAAGUGGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCGCCGCUGGAAGCG______AUUGGCUGGCGUGUGAAUCCC ......((((((....(((.((((((((((....))))....)))))).)))..........(..(((..(.......(((.....))).......)..)))..)))))))..... (-32.43 = -32.99 + 0.56)


| Location | 16,746,049 – 16,746,163 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -50.70 |
| Consensus MFE | -41.06 |
| Energy contribution | -40.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16746049 114 + 22224390 GGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCGCCGCUGGAAGCG------AUUGGCUGGCGUGUGAAUCCCACAUGCUGCGCAGUCGACUUCCGCACAGUGGGCCCAAAUU (((((((((.........((((....))))(((....)))))))).(((((((------((((((.((((((((.....)))))))))).)))))).))))).........))))..... ( -51.80) >DroSec_CAF1 33390 120 + 1 GGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCGCCGCUGGAAGCGGUUUGGAUUGGCUGGCGUGUGAAUCCCACAUGCUGCGCAGUCGACUUCCGCACAGUGGGCCCAAAUU ((((....(((..((.....))..)))...(((....))).((((((...((((..(.(((((((.((((((((.....)))))))))).))))).)...)))).))))))))))..... ( -50.90) >DroSim_CAF1 31724 114 + 1 GGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCGCCGCUGGAAGCG------AUUGGCUGGCGUGUGAAUCCCACAUGCUGCGCAGUCGACUUCCGCACAGUGGGCCCAAAUU (((((((((.........((((....))))(((....)))))))).(((((((------((((((.((((((((.....)))))))))).)))))).))))).........))))..... ( -51.80) >DroEre_CAF1 25046 114 + 1 GGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCACCGCUGGAAGCG------AUUGGCUGGCGUGUGAAUCCCACAUGCUGCGCAGUCGACUUCCGCACAGUGGGCCCAAGUU (((((((((((..((.....))..)))...(((....))).)))).(((((((------((((((.((((((((.....)))))))))).)))))).))))).........))))..... ( -49.60) >DroYak_CAF1 24843 114 + 1 GGGCGUGGCAUCAGAUAAUUUCUAAUGAAAGUUGAAAAGAGCCGCUGGAAGCG------AUUGGCUGGCGUGUGAAUCCCACAUGCUGCGCAGUCGACUUCCGCACAGUGGGCCCAAGUU (((((((((.((((....((((....)))).)))).....))))).(((((((------((((((.((((((((.....)))))))))).)))))).))))).........))))..... ( -49.40) >consensus GGGCGUGGCAUCGGAUAAUUUCUAAUGAAAGUUGAAAAGCGCCGCUGGAAGCG______AUUGGCUGGCGUGUGAAUCCCACAUGCUGCGCAGUCGACUUCCGCACAGUGGGCCCAAAUU ((((......((((....((((....)))).))))......((((((...(((......((((((.((((((((.....)))))))))).)))).......))).))))))))))..... (-41.06 = -40.90 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:47 2006