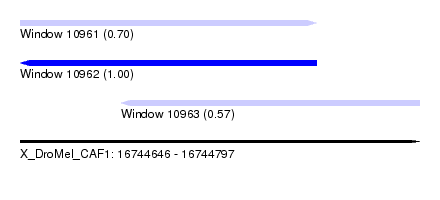
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,744,646 – 16,744,797 |
| Length | 151 |
| Max. P | 0.998101 |

| Location | 16,744,646 – 16,744,758 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -18.51 |
| Consensus MFE | -3.73 |
| Energy contribution | -3.89 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16744646 112 + 22224390 AAUAAAUAAAUCGACAAUUAAGCGAGGGACA--GUCGGCGGGUACUGCCACCCCACUUCCCU-CGUUUCCAUCC----UAUCUAUGGAUAUGAAUAUACAUACAUAUAUUUAUCCA-CAU ...................((((((((((.(--((.((..((.....))..)).))))))))-)))))......----......((((((..((((((......))))))))))))-... ( -30.90) >DroSec_CAF1 31996 91 + 1 AAUAAAUAAAUCGACAAUUAAGCGAGGGACA--GCA---CGGCACUGGCACCCCACUUCCCC--------CUCC----CAUCUAUAGAUAU------------AUGUAUAUAUCCACCAU .....................(.(((((...--((.---..))..(((....))).....))--------))).----).......(((((------------(....))))))...... ( -12.40) >DroSim_CAF1 30324 91 + 1 AAUAAAUAAAUCGACAAUUAAGCGAGGGACA--GCA---CGGCACUUGCACCCCACUUCCCC--------CUCC----CAUCUAUAGAUAU------------AUGUAUAUAUCCACCAU .....................(.(((((..(--(..---..((....))......))...))--------))).----).......(((((------------(....))))))...... ( -11.70) >DroEre_CAF1 23455 96 + 1 AAUAAAUAAGACGACAAUUAAACGAGGGACA--GCU---UGGAACUGCCACCCCACUCCUCC--GAUUCCUUCC----UAUCUAUGGAUGUGGAU--------AU----UUAUCCA-CAU ......................((..(((..--...---(((.....)))......)))..)--)......(((----.......)))(((((((--------..----..)))))-)). ( -18.50) >DroYak_CAF1 23475 108 + 1 AAUAAAUAAAACGACAAUUAAACGAGGGACAACGCU---CGCUGCUGCCACCCCACUCCUCCCCAUUUCCCUUCAAUGUAUCUAUGGAUGUGGAU--------AUACAUAUAUCCA-CAU .......................(((((........---.((....))........))))).((((.................))))((((((((--------((....)))))))-))) ( -19.06) >consensus AAUAAAUAAAUCGACAAUUAAGCGAGGGACA__GCA___CGGCACUGCCACCCCACUUCCCC____UUCCCUCC____UAUCUAUGGAUAUG_AU________AUAUAUAUAUCCA_CAU .........................(((.......................)))..............................((((((....................)))))).... ( -3.73 = -3.89 + 0.16)


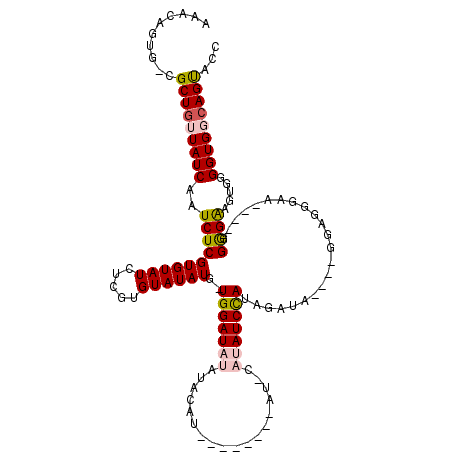
| Location | 16,744,646 – 16,744,758 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -12.66 |
| Energy contribution | -12.46 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.40 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16744646 112 - 22224390 AUG-UGGAUAAAUAUAUGUAUGUAUAUUCAUAUCCAUAGAUA----GGAUGGAAACG-AGGGAAGUGGGGUGGCAGUACCCGCCGAC--UGUCCCUCGCUUAAUUGUCGAUUUAUUUAUU .((-(((((((((((((....)))))))..))))))))((((----(...((...((-((((((((.((.(((......))))).))--).)))))))))...)))))............ ( -36.50) >DroSec_CAF1 31996 91 - 1 AUGGUGGAUAUAUACAU------------AUAUCUAUAGAUG----GGAG--------GGGGAAGUGGGGUGCCAGUGCCG---UGC--UGUCCCUCGCUUAAUUGUCGAUUUAUUUAUU ...(((((((((....)------------))))))))...((----((.(--------(((((((((.((((....)))).---)))--).)))))).)))).................. ( -27.70) >DroSim_CAF1 30324 91 - 1 AUGGUGGAUAUAUACAU------------AUAUCUAUAGAUG----GGAG--------GGGGAAGUGGGGUGCAAGUGCCG---UGC--UGUCCCUCGCUUAAUUGUCGAUUUAUUUAUU ...(((((((((....)------------))))))))...((----((.(--------(((((((((.((((....)))).---)))--).)))))).)))).................. ( -27.70) >DroEre_CAF1 23455 96 - 1 AUG-UGGAUAA----AU--------AUCCACAUCCAUAGAUA----GGAAGGAAUC--GGAGGAGUGGGGUGGCAGUUCCA---AGC--UGUCCCUCGUUUAAUUGUCGUCUUAUUUAUU (((-(((((..----..--------))))))))..(((((((----(((.(((((.--.((.((..((((..((.......---.))--..)))))).))..))).)).)))))))))). ( -32.80) >DroYak_CAF1 23475 108 - 1 AUG-UGGAUAUAUGUAU--------AUCCACAUCCAUAGAUACAUUGAAGGGAAAUGGGGAGGAGUGGGGUGGCAGCAGCG---AGCGUUGUCCCUCGUUUAAUUGUCGUUUUAUUUAUU (((-(((((((....))--------))))))))............((((..(((((((.((.((((((((.((((((....---...))))))))))))))..)).))))))).)))).. ( -31.90) >consensus AUG_UGGAUAUAUACAU________AU_CAUAUCCAUAGAUA____GGAGGGAA____GGGGAAGUGGGGUGGCAGUACCG___AGC__UGUCCCUCGCUUAAUUGUCGAUUUAUUUAUU ......((((.....................(((....))).....................((((((((.((((((........))..))))))))))))...))))............ (-12.66 = -12.46 + -0.20)



| Location | 16,744,684 – 16,744,797 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16744684 113 - 22224390 AAACAGUG-CGCUGUUAUCAAUCUCGUGUAUCUCGUGUAUAUG-UGGAUAAAUAUAUGUAUGUAUAUUCAUAUCCAUAGAUA----GGAUGGAAACG-AGGGAAGUGGGGUGGCAGUACC ........-.(((((((((..((((((...((((.(((...((-(((((((((((((....)))))))..)))))))).)))----.)).))..)))-))).......)))))))))... ( -33.40) >DroSec_CAF1 32031 95 - 1 AAACAGUG-CGCUGUUAUCAAUCUCGUGUAUCUCGUGUAUAUGGUGGAUAUAUACAU------------AUAUCUAUAGAUG----GGAG--------GGGGAAGUGGGGUGCCAGUGCC .......(-(((((.((((..((((.(...((((((.(((.....(((((((....)------------))))))))).)))----))))--------.)))).....)))).)))))). ( -25.20) >DroSim_CAF1 30359 95 - 1 AAACAGUG-CGCUGUUAUCAAUCUCGUGUAUCUCGUGUAUAUGGUGGAUAUAUACAU------------AUAUCUAUAGAUG----GGAG--------GGGGAAGUGGGGUGCAAGUGCC .....(..-(.(..((.((..((((((.(((...(((((((((.....)))))))))------------......))).)))----))).--------.))..))..).)..)....... ( -24.50) >DroEre_CAF1 23490 101 - 1 AAACAGUGUCGCUGUUAUCAAUCUCGUGUAUCUCAUGUAUAUG-UGGAUAA----AU--------AUCCACAUCCAUAGAUA----GGAAGGAAUC--GGAGGAGUGGGGUGGCAGUUCC ..........(((((((((...(((...(((((.(((...(((-(((((..----..--------)))))))).))))))))----.((.....))--....)))...)))))))))... ( -27.60) >DroYak_CAF1 23512 111 - 1 AAACAGUGUCGCUGUUAUCAAUCUCGUGUAUCUCAUGUAUAUG-UGGAUAUAUGUAU--------AUCCACAUCCAUAGAUACAUUGAAGGGAAAUGGGGAGGAGUGGGGUGGCAGCAGC ..........(((((((((...(((((((((((.(((...(((-(((((((....))--------)))))))).))))))))))).................)))...)))))))))... ( -36.32) >consensus AAACAGUG_CGCUGUUAUCAAUCUCGUGUAUCUCGUGUAUAUG_UGGAUAUAUACAU________AU_CAUAUCCAUAGAUA____GGAGGGAA____GGGGAAGUGGGGUGGCAGUACC ..........(((((((((..((((((((((.....))))))..(((((((..................))))))).......................)))).....)))))))))... (-15.87 = -16.39 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:43 2006