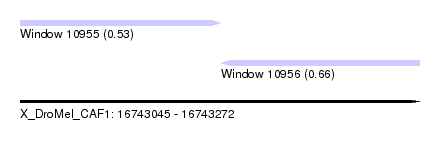
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,743,045 – 16,743,272 |
| Length | 227 |
| Max. P | 0.660139 |

| Location | 16,743,045 – 16,743,159 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -25.86 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
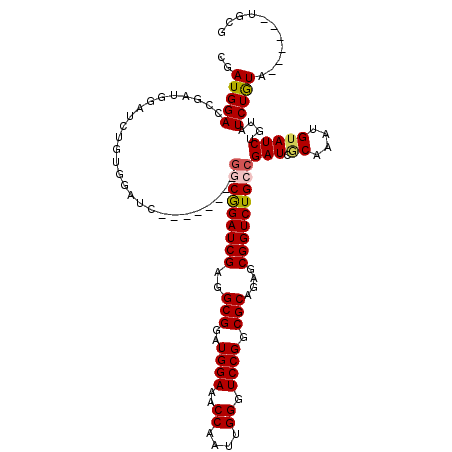
>X_DroMel_CAF1 16743045 114 + 22224390 CAAUGGACCGAUGGAUCUGUGGAUCUCUGGAUCGGCGGAUCGAGGCGGAUGGAAACCAAUUGGGUCCGGCGCAGAGCGGUCUGCCGAUCUCAAAUGUAUCUGUAUCUGUU------UGCG ....((((.((((((((....))))..(((((((((((((((..(((..((((..((....)).)))).)))....))))))))))))).))..........)))).)))------)... ( -43.40) >DroSec_CAF1 30490 111 + 1 CAAUGGACCGAUGGAUCUGUGGAUCUCU-----GGCGGAUCGAGGCGGAUGGAAACCAAUUGGGUCCGGCGCAGAGCGGUCUGCCGAUCGCAAAUGUAUCUGUAUCUGUAUC----UGUG ......((.((((.((.((((.(((...-----(((((((((..(((..((((..((....)).)))).)))....)))))))))))))))).)).)))).)).........----.... ( -37.00) >DroSim_CAF1 28720 102 + 1 CGAUGGACCGAUGGAGCUCUGGAUC--------GGCGGAUCGAGGCGGAUGGAAACCAAUUGGGUCCGGCGCAGAGCGGUCUGCCGAUCGCAAAUGUAUCUGUAUCUGUA---------- ..(((((...(((((((..((((((--------(((((((((..(((..((((..((....)).)))).)))....))))))))))))).))...)).))))).))))).---------- ( -41.10) >DroEre_CAF1 21898 112 + 1 CGAUGGAUCGGCGGAUCGGCGGAUC--------GGCGGAUCGAGGCGGAUGGAAACCAAUUGGGUCCGGCGCAGAGCGGUCUGGCGAUCACAAAUGUAUCUGUAUCUGUAUCUGUGUGCG .((((.(...((((((((...((((--------(.(((((((..(((..((((..((....)).)))).)))....))))))).))))).....)).))))))...).))))........ ( -40.90) >DroYak_CAF1 21898 90 + 1 CGAUGGAUCGG------------------------CAGAUCGAGGCGGAUGGAAACCAAUUGGGUCCGGCGCAGAGCGGUCUGCCGAUCGCAGAUGUAUCUGUAUCUAU------AUGCG .(((.((((((------------------------(((((((..(((..((((..((....)).)))).)))....)))))))))))))(((((....))))))))...------..... ( -41.20) >consensus CGAUGGACCGAUGGAUCUGUGGAUC________GGCGGAUCGAGGCGGAUGGAAACCAAUUGGGUCCGGCGCAGAGCGGUCUGCCGAUCGCAAAUGUAUCUGUAUCUGUA______UGCG ..(((((..........................(((((((((..(((..((((..((....)).)))).)))....)))))))))(((.((....)))))....)))))........... (-25.86 = -26.22 + 0.36)

| Location | 16,743,159 – 16,743,272 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
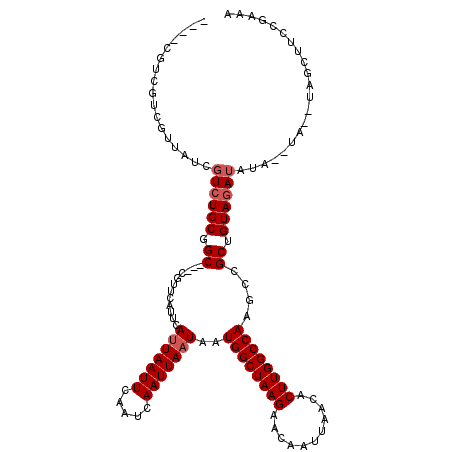
>X_DroMel_CAF1 16743159 113 - 22224390 ACUACGUCGUCGUUAUCGUCUGCCGC---CGUUCAUUCAUUAAUUCAAUCAAUUAAUAAUGGCUAAGAACAAUUAACACUUGGCCAAACCGCUGUAUAU----AUACAUAGCUUCCUACA .....((((..((((...(((...((---((((.....(((((((.....)))))))))))))..))).....))))...))))......(((((....----....)))))........ ( -18.50) >DroSec_CAF1 30601 110 - 1 -------CGUCGUUAUCGUCUGCGGC---CGUUCAUUCAUUAAUUCAAUCAAUUAAUAAUGGCUAAGAACAAUUAACACUUGGCCAAGUCGCUGUAGAUAUACAUACAUAGCUUCCGAGA -------..........(((((((((---.........(((((((.....)))))))..((((((((...........))))))))....)))))))))..................... ( -24.50) >DroSim_CAF1 28822 88 - 1 -------CGUCGUUAUCGUCUGCGGC---CGUUCAUUCAUUAAUUCAAUCAAUUAAUAAUGGCUAAGAACAAUUAACACUUGGCCAAGCCGCUGUAGA---------------------- -------...........((((((((---.(((.....(((((((.....)))))))..((((((((...........))))))))))).))))))))---------------------- ( -23.80) >DroEre_CAF1 22010 108 - 1 UCAUCGUUGUCGUUGUCGUCUGCAGC---CGUUCAUUCAUUAAUUCAAUCAAUUAAUAAUGGCUAAGAACAAUUAACACUUGGCCAAGCCGCAGUAGAUAUA--------GC-UCCAAAA ......(((..(((((.((((((.((---.(((.....(((((((.....)))))))..((((((((...........))))))))))).)).)))))))))--------))-..))).. ( -26.90) >DroYak_CAF1 21988 120 - 1 AGUUCGUCGUCGUUAUCGUCUGCUGCCGUCGUUCAUUCAUUAAUUCAAUCAAUUAUUAAUGGCUAAGAACAAUUAACACUUGGCCAAGCCGCUGUAGAUAUAUGUAUGUAGCUUCCGAAA (((((((...(((....((((((.((.((.........((((((..........))))))(((((((...........)))))))..)).)).))))))..))).))).))))....... ( -21.50) >consensus ____CGUCGUCGUUAUCGUCUGCGGC___CGUUCAUUCAUUAAUUCAAUCAAUUAAUAAUGGCUAAGAACAAUUAACACUUGGCCAAGCCGCUGUAGAUAUA__UA__UAGCUUCCGAAA .................((((((.((............(((((((.....)))))))..((((((((...........))))))))....)).))))))..................... (-17.30 = -17.90 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:37 2006