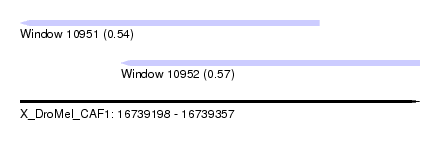
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,739,198 – 16,739,357 |
| Length | 159 |
| Max. P | 0.565760 |

| Location | 16,739,198 – 16,739,317 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.80 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16739198 119 - 22224390 UCAAUUGCAAUUUUUAUAACGCUGCCCAGCGUUUUUAUGCCGCAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG-GUUUUUCUCUCUUAUCGCAAGGUCGUUCAUAAAAAACAUAUAU ..........(((((((((((..(((..(((.......((((.((((((...((((....))))...)))))).))-)).............)))..))))))).)))))))........ ( -25.25) >DroSec_CAF1 26882 118 - 1 UGAAUUGCAAUUUUUAUAACGCUGACCAGCGUUUUUAUGCCGCAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG-GUUUUUCUCUCUUAUCGCAAGGUCGUUCAUAAA-AACAUAUAU .((.((((.........((((((....)))))).....((((.((((((...((((....))))...)))))).))-))..............))))..)).........-......... ( -26.30) >DroSim_CAF1 25022 101 - 1 UCAAUUGCAAUUUUUAUAACGCUGACCAGCGUUUUUAUGCCGCAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG-GUUUUUCUCUCUUAUCGCAAGGUCG------------------ ....((((.........((((((....)))))).....((((.((((((...((((....))))...)))))).))-))..............)))).....------------------ ( -24.70) >DroEre_CAF1 18192 118 - 1 UCAAUUGCAAUUUUUAUAACGCUGCCCAGCGUUUUUAUGCCACAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG-UUUUUUCUCUCUUAUCGCAAGGCCGUUUAUAAA-AACAUAUAU ..........(((((((((.((.(((..(((.....................((((....))))....((((....-...))))........)))..))).)))))))))-))....... ( -20.10) >DroYak_CAF1 18254 119 - 1 UCAAUUGCAAUUUUUAUAACGCUGUCCAGCGUUUUUAUGCCACAUUUUAUUGCUUAACAAUGGGAUCUGAAAUUCGGUUUUUUCUCUCUUAUCGCAAGGCCGUUCAUGAA-AACAAAAAC ....................(((....)))(((((((((.((.(((((((((.....))))))))).)).....(((((((..............)))))))..))))))-)))...... ( -21.34) >consensus UCAAUUGCAAUUUUUAUAACGCUGACCAGCGUUUUUAUGCCGCAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG_GUUUUUCUCUCUUAUCGCAAGGUCGUUCAUAAA_AACAUAUAU ....((((.........((((((....)))))).......((.((((((...((((....))))...)))))).)).................))))....................... (-17.36 = -17.44 + 0.08)



| Location | 16,739,238 – 16,739,357 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16739238 119 - 22224390 ACCCCGAAAACAAGAAUGUCGCUUUUUGCUCGGAUCCAUUUCAAUUGCAAUUUUUAUAACGCUGCCCAGCGUUUUUAUGCCGCAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG-GUU ...(((((....(((((((.((...((((...((......))....)))).......((((((....)))))).....)).)))))))....((((....))))........))))-).. ( -28.80) >DroSec_CAF1 26921 119 - 1 ACCCCGAAAACAAGAAUGUCGCUUUUUGCUCGGAUCCAUUUGAAUUGCAAUUUUUAUAACGCUGACCAGCGUUUUUAUGCCGCAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG-GUU ...(((((....(((((((.((...(((((((((....)))))...)))).......((((((....)))))).....)).)))))))....((((....))))........))))-).. ( -28.20) >DroSim_CAF1 25044 119 - 1 ACCCCGAAAACAAGAAUGUCGCUUUUUGCUCGGAUCCAUUUCAAUUGCAAUUUUUAUAACGCUGACCAGCGUUUUUAUGCCGCAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG-GUU ...(((((....(((((((.((...((((...((......))....)))).......((((((....)))))).....)).)))))))....((((....))))........))))-).. ( -28.80) >DroEre_CAF1 18231 119 - 1 ACCCCGAAAACGAGAAUGUCGCUUUUUGCUCGGAUCCAUUUCAAUUGCAAUUUUUAUAACGCUGCCCAGCGUUUUUAUGCCACAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG-UUU .......((((((((((((.((...((((...((......))....)))).......((((((....)))))).....)).)))))).....((((....)))).........)))-))) ( -25.90) >DroYak_CAF1 18293 120 - 1 ACCCCGAAGACAAGAAUGCCACUUUUUGCUCGGAUCCAUUUCAAUUGCAAUUUUUAUAACGCUGUCCAGCGUUUUUAUGCCACAUUUUAUUGCUUAACAAUGGGAUCUGAAAUUCGGUUU ...((((((.((((((......)))))))((((((((.........(((((......((((((....))))))...............))))).........))))))))..)))))... ( -29.57) >consensus ACCCCGAAAACAAGAAUGUCGCUUUUUGCUCGGAUCCAUUUCAAUUGCAAUUUUUAUAACGCUGACCAGCGUUUUUAUGCCGCAUUUUAUUGCCCAACAAUGGGAACUGAAAUUCG_GUU ....((((....(((((((.((...((((...((......))....)))).......((((((....)))))).....)).)))))))....((((....))))........)))).... (-21.76 = -21.92 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:33 2006