
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,736,603 – 16,736,817 |
| Length | 214 |
| Max. P | 0.944130 |

| Location | 16,736,603 – 16,736,704 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.02 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -17.18 |
| Energy contribution | -16.14 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16736603 101 + 22224390 UUGUAUUAUCAGCAUGUG-------------------UCUACAUACCAGGCUCUGGGUCUUUGAGUUUAAUGUAUUCAAACAUUGAACUUUUAUUUACUGUGCAAGUGACCAUAAUGUUG .........(((((((.(-------------------(((.......)))).)..((((...(((((((((((......)))))))))))......(((.....)))))))...)))))) ( -23.10) >DroSec_CAF1 24248 94 + 1 -----CUAUCAGCAUGUG-------------------UCUACAUACCAGGC--UGGGUCUUUGAGUUUAAUGUAUUCAAACAUUGAACUUUUAUUUACUGUGCAAGUGACCAUAAUGCUG -----....((((((..(-------------------(((.......))))--..((((...(((((((((((......)))))))))))......(((.....)))))))...)))))) ( -25.20) >DroSim_CAF1 22617 94 + 1 -----CUAUCAGCAUGUG-------------------UCUACAUACCAGGC--UGGGUCUUUGAGUUUAAUGUAUUCAAACAUUGAACUUUUAUUUACUGUGCAAGUGACCAUAAUGCUG -----....((((((..(-------------------(((.......))))--..((((...(((((((((((......)))))))))))......(((.....)))))))...)))))) ( -25.20) >DroEre_CAF1 15623 105 + 1 -----UUAUCAGUACAUAUCUACUAUAUCUAC-----UAUACAUGCCAGGC--UGGGUCUUUAAGUUUAAUGUAUUCAAACAUUGAACUUUUAUUUGCUGUGCAAGUGACCAUAAU---G -----.....((((.((((.....)))).)))-----).............--..((((...(((((((((((......)))))))))))..((((((...)))))))))).....---. ( -21.20) >DroYak_CAF1 15652 109 + 1 -----UUAUCAGCACAUA-CUCGUAUAAGUAUAAGUAUCUACAUACUAGGC--UGGGUCUUUAAGUUUAAUGUAUUCAAACAUUGAACUUUUAUUUACUGUGCAAGUGACCAUAAU---G -----....((((..(((-((......))))).(((((....)))))..))--))((((...(((((((((((......)))))))))))......(((.....))))))).....---. ( -26.10) >consensus _____UUAUCAGCAUGUG___________________UCUACAUACCAGGC__UGGGUCUUUGAGUUUAAUGUAUUCAAACAUUGAACUUUUAUUUACUGUGCAAGUGACCAUAAUG_UG ...........(((((.............................(((.....)))......(((((((((((......)))))))))))........)))))................. (-17.18 = -16.14 + -1.04)

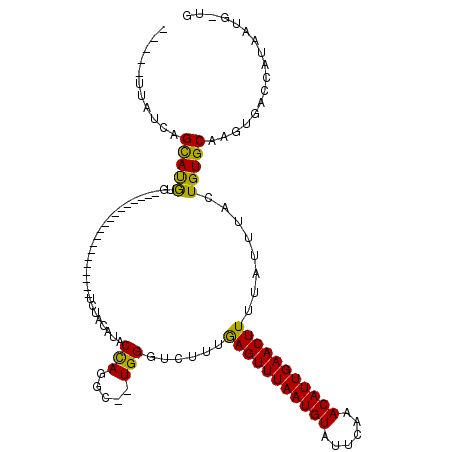

| Location | 16,736,603 – 16,736,704 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.02 |
| Mean single sequence MFE | -20.18 |
| Consensus MFE | -13.72 |
| Energy contribution | -13.12 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16736603 101 - 22224390 CAACAUUAUGGUCACUUGCACAGUAAAUAAAAGUUCAAUGUUUGAAUACAUUAAACUCAAAGACCCAGAGCCUGGUAUGUAGA-------------------CACAUGCUGAUAAUACAA ....((((((((((((.....))).......((((.(((((......))))).))))....))))........(((((((...-------------------.))))))).))))).... ( -17.40) >DroSec_CAF1 24248 94 - 1 CAGCAUUAUGGUCACUUGCACAGUAAAUAAAAGUUCAAUGUUUGAAUACAUUAAACUCAAAGACCCA--GCCUGGUAUGUAGA-------------------CACAUGCUGAUAG----- ((((((....(((((.(((.(((........((((.(((((......))))).))))..........--..)))))).)).))-------------------)..))))))....----- ( -19.75) >DroSim_CAF1 22617 94 - 1 CAGCAUUAUGGUCACUUGCACAGUAAAUAAAAGUUCAAUGUUUGAAUACAUUAAACUCAAAGACCCA--GCCUGGUAUGUAGA-------------------CACAUGCUGAUAG----- ((((((....(((((.(((.(((........((((.(((((......))))).))))..........--..)))))).)).))-------------------)..))))))....----- ( -19.75) >DroEre_CAF1 15623 105 - 1 C---AUUAUGGUCACUUGCACAGCAAAUAAAAGUUCAAUGUUUGAAUACAUUAAACUUAAAGACCCA--GCCUGGCAUGUAUA-----GUAGAUAUAGUAGAUAUGUACUGAUAA----- .---.....((((..((((...))))....(((((.(((((......))))).)))))...))))((--(....((((((.((-----.((....)).)).)))))).)))....----- ( -21.30) >DroYak_CAF1 15652 109 - 1 C---AUUAUGGUCACUUGCACAGUAAAUAAAAGUUCAAUGUUUGAAUACAUUAAACUUAAAGACCCA--GCCUAGUAUGUAGAUACUUAUACUUAUACGAG-UAUGUGCUGAUAA----- .---.....(((((((.....)))......(((((.(((((......))))).)))))...))))((--((..(((((....)))))(((((((....)))-)))).))))....----- ( -22.70) >consensus CA_CAUUAUGGUCACUUGCACAGUAAAUAAAAGUUCAAUGUUUGAAUACAUUAAACUCAAAGACCCA__GCCUGGUAUGUAGA___________________CACAUGCUGAUAA_____ .........((((..((((...)))).....((((.(((((......))))).))))....))))........(((((((.......................))))))).......... (-13.72 = -13.12 + -0.60)



| Location | 16,736,704 – 16,736,817 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.52 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -20.91 |
| Energy contribution | -20.83 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16736704 113 - 22224390 GGUUUUGCAUAAAAGUACAAAAAGCAUUCGUGUUUACAAAUCAAUAAUGCAAAAAUCCGCACAGUGGGCGUAAUCAACAACCGUUGC-------GACCACGCCCUCUCCUUCCUCAUCAU ((((((((((...........(((((....)))))...........))))))))..))....((.((((((...((((....)))).-------....)))))).))............. ( -23.45) >DroSec_CAF1 24342 113 - 1 GGUUUUGCAUAAAUGUACAAAAAGCAUUCGUGUUUACAAAUCAAUAAUGCAAAAAUCCGCACGGUGGGCGUAAUCAACAACCGUUGC-------GACCACGCCCUCCCCUUCCUCAUCAU ((((((((((.(.((......(((((....)))))......)).).))))))))..))....((.((((((...((((....)))).-------....)))))).))............. ( -26.60) >DroSim_CAF1 22711 113 - 1 GGUUUUGCAUAAAUGUACAAAAAGCAUUCGUGUUUACAAAUCAAUAAUGCAAAAAUCCGCACGGUGGGCGUAAUCAACAACCGUUGC-------GACCACGCCCUCCCCUUCCUCAUCAU ((((((((((.(.((......(((((....)))))......)).).))))))))..))....((.((((((...((((....)))).-------....)))))).))............. ( -26.60) >DroEre_CAF1 15728 111 - 1 GGUUUUGCAUAAAAGUACAAAGAGCACUAGUGUUUACAAAUCAAUAAUGCAAAAAUCCGCAUG-UGGGCGUAAUCAACAACCGUUGC-------GACCACGCCCUCC-CUUCCUCAUCAU ((((((((((...........(((((....)))))...........))))))))..)).....-.((((((...((((....)))).-------....))))))...-............ ( -23.65) >DroYak_CAF1 15761 120 - 1 GGUUUUGCAUAAAAGUACAAAAAGCAUUUGUGUUUACAAAUCAAUAAUGCAAAAAUCCGCAUGGUAGGCGUAAUCAACAACCGUUGCUGUUGUUGACCACGCCCUCCCCUUCCUCAUCAU (((((.(((((((.((.......)).)))))))....)))))....((((........))))((..(((((..((((((((.......))))))))..)))))..))............. ( -31.00) >consensus GGUUUUGCAUAAAAGUACAAAAAGCAUUCGUGUUUACAAAUCAAUAAUGCAAAAAUCCGCACGGUGGGCGUAAUCAACAACCGUUGC_______GACCACGCCCUCCCCUUCCUCAUCAU ((((((((((...........(((((....)))))...........))))))))..))....((.((((((...((((....))))............)))))).))............. (-20.91 = -20.83 + -0.08)

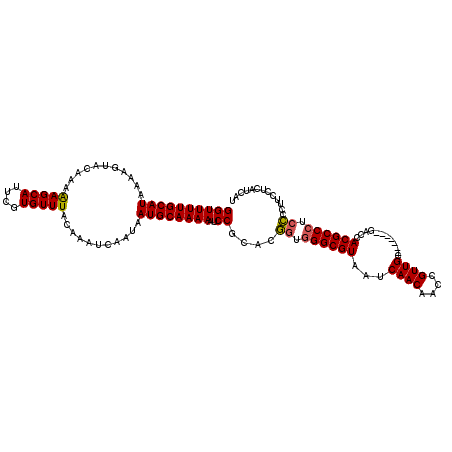

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:30 2006