| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,868,982 – 1,869,088 |
| Length | 106 |
| Max. P | 0.654139 |

| Location | 1,868,982 – 1,869,088 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.55 |
| Mean single sequence MFE | -37.54 |
| Consensus MFE | -25.23 |
| Energy contribution | -26.19 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
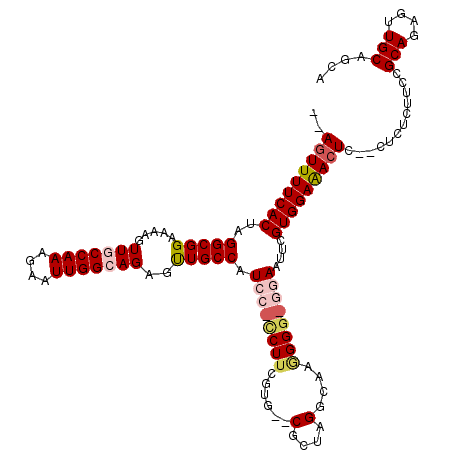
>X_DroMel_CAF1 1868982 106 + 22224390 UGCUGCAACUCUGCGGAA--------ACUCUCCACGAAUUCC-CCCCUUGCCUAGCG--CACGAAGA-GGAUGGCAGCUCUGCCAAUUCUUUGGUAACUUUUCCGCCUAGUGAAAACU-- .......(((..((((((--------(...............-.....(((.....)--))((((((-(..(((((....))))).)))))))......)))))))..))).......-- ( -27.40) >DroSec_CAF1 6291 112 + 1 UGCUGCAGCUCUGCGGAAGAGAG--GAGUUUCCACGAAUUCC-CCCCUUGCCUAGCG--CACGAAGG-GGAUGGCAACUCUGCCAAUUCUUUGGCAACUUUUCCGCCUAGUGAAAACU-- .......(((..(((((((((.(--(((((.(((.....(((-((.(.(((.....)--)).)..))-)))))).))))))(((((....)))))..)))))))))..))).......-- ( -42.80) >DroSim_CAF1 1533 112 + 1 UGCUGCAGCUCUGCGGAAGAGAG--GAGUUUCCACGAAUUCC-CCCCUUGCCUAGCG--CACGAAGG-GGAUGGCAACUCUGCCAAUUCUUUGGCAACUUUUCCGCCUAGUGAAAACU-- .......(((..(((((((((.(--(((((.(((.....(((-((.(.(((.....)--)).)..))-)))))).))))))(((((....)))))..)))))))))..))).......-- ( -42.80) >DroEre_CAF1 4962 116 + 1 UGCUGCAACUCUGCGGAAGAGAGUAGAGUUUCCACGGAUUU--CCCUUUGCCUUGCG--CCCGAAGUGUGAUGGCAACUCUUCCAAUUCUUUGGCAACUUUUCCGCCUAGUGAAAACUGU ............(((((((((...((((((.....((....--))....((((..((--(.....)))..).))))))))).((((....))))...))))))))).((((....)))). ( -34.90) >DroYak_CAF1 6914 120 + 1 UGUUGCAACUCUGCGGAAGAGAGUAGAGUUUCCACGGAUUUUCCCCUUUGGCUUGCGCGCACGUAGGAUGAUGGCAACUCUGCCAAUUCUUUGGCAAUUUUUCCGCCUAGUGAAAACUGU .(((...(((..(((((((((...((((((.(((.((.....)).......((((((....))))))....))).))))))(((((....)))))..)))))))))..)))...)))... ( -39.80) >consensus UGCUGCAACUCUGCGGAAGAGAG__GAGUUUCCACGAAUUCC_CCCCUUGCCUAGCG__CACGAAGG_GGAUGGCAACUCUGCCAAUUCUUUGGCAACUUUUCCGCCUAGUGAAAACU__ .......(((..(((((((((....(((((.(((..............(((...)))..............))).)))))((((((....)))))).)))))))))..)))......... (-25.23 = -26.19 + 0.96)


| Location | 1,868,982 – 1,869,088 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.55 |
| Mean single sequence MFE | -39.46 |
| Consensus MFE | -25.05 |
| Energy contribution | -26.64 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1868982 106 - 22224390 --AGUUUUCACUAGGCGGAAAAGUUACCAAAGAAUUGGCAGAGCUGCCAUCC-UCUUCGUG--CGCUAGGCAAGGGG-GGAAUUCGUGGAGAGU--------UUCCGCAGAGUUGCAGCA --.(((.(.(((..(((((((..((.(((..(((((((((....)))))(((-((((..((--(.....))).))))-))))))).)))))..)--------))))))..))).).))). ( -38.40) >DroSec_CAF1 6291 112 - 1 --AGUUUUCACUAGGCGGAAAAGUUGCCAAAGAAUUGGCAGAGUUGCCAUCC-CCUUCGUG--CGCUAGGCAAGGGG-GGAAUUCGUGGAAACUC--CUCUCUUCCGCAGAGCUGCAGCA --.(((..((((..((((((.((.((((((....))))))(((((.((((((-((((..((--(.....))).))))-))).....))).)))))--..)).))))))..)).)).))). ( -41.70) >DroSim_CAF1 1533 112 - 1 --AGUUUUCACUAGGCGGAAAAGUUGCCAAAGAAUUGGCAGAGUUGCCAUCC-CCUUCGUG--CGCUAGGCAAGGGG-GGAAUUCGUGGAAACUC--CUCUCUUCCGCAGAGCUGCAGCA --.(((..((((..((((((.((.((((((....))))))(((((.((((((-((((..((--(.....))).))))-))).....))).)))))--..)).))))))..)).)).))). ( -41.70) >DroEre_CAF1 4962 116 - 1 ACAGUUUUCACUAGGCGGAAAAGUUGCCAAAGAAUUGGAAGAGUUGCCAUCACACUUCGGG--CGCAAGGCAAAGGG--AAAUCCGUGGAAACUCUACUCUCUUCCGCAGAGUUGCAGCA ...(((.(.(((..((((((.((...((((....)))).((((((.((((....(((..(.--(....).).)))((--....)))))).))))))...)).))))))..))).).))). ( -33.90) >DroYak_CAF1 6914 120 - 1 ACAGUUUUCACUAGGCGGAAAAAUUGCCAAAGAAUUGGCAGAGUUGCCAUCAUCCUACGUGCGCGCAAGCCAAAGGGGAAAAUCCGUGGAAACUCUACUCUCUUCCGCAGAGUUGCAACA ...(((.(.(((..((((((.....(((((....)))))((((((.((((..((((...((.((....))))...))))......)))).))))))......))))))..))).).))). ( -41.60) >consensus __AGUUUUCACUAGGCGGAAAAGUUGCCAAAGAAUUGGCAGAGUUGCCAUCC_CCUUCGUG__CGCUAGGCAAGGGG_GGAAUUCGUGGAAACUC__CUCUCUUCCGCAGAGUUGCAGCA ..(((((((((..(((((.....(((((((....)))))))..))))).((((((((......(....)....))))))))....)))))))))............(((....))).... (-25.05 = -26.64 + 1.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:48 2006