
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,733,068 – 16,733,201 |
| Length | 133 |
| Max. P | 0.999473 |

| Location | 16,733,068 – 16,733,181 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -48.98 |
| Consensus MFE | -37.38 |
| Energy contribution | -38.08 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.35 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
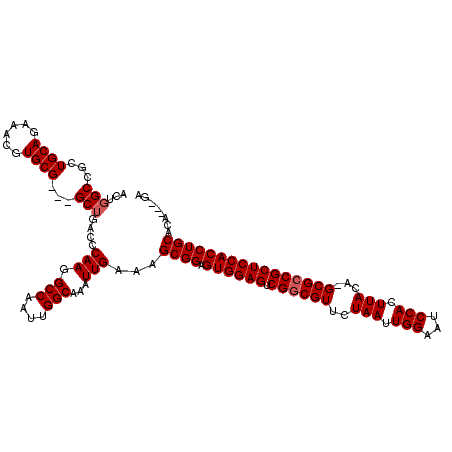
>X_DroMel_CAF1 16733068 113 + 22224390 ACUGGCCGCUGCAGAAACGUGCG---GCUGACCCAAGGCCAAUUGGCAAAUUGAAAGCGGAGUGGAGUCGUCGUUCUAAUUGGAAUCCACUUACA-GCGCCGCUCCACCUGCACA---GA .((((((((.((......)))))---)).....(((.(((....)))...)))...((((.((((((.((.((((.(((.(((...))).))).)-))).)))))))))))).))---). ( -44.60) >DroSec_CAF1 20840 88 + 1 ----------------------------UGACCCAAGGCCAAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACA-GCGCCGCUCCACCUGCACA---GA ----------------------------.....(((.(((....)))...)))...((((.((((((.(((((((.(((.(((...))).))).)-))))))))))))))))...---.. ( -39.20) >DroSim_CAF1 18198 113 + 1 ACUGGCCGCUGCAGAAACGUGCG---GCUGACCCAAGGCCAAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACA-GCGCCGCUCCACCUGCACA---GA .((((((((.((......)))))---)).....(((.(((....)))...)))...((((.((((((.(((((((.(((.(((...))).))).)-)))))))))))))))).))---). ( -51.70) >DroEre_CAF1 12166 117 + 1 ACUGGCCGCUGCAGAAACGUGCGGCGGCUGACCCAAGGCCAAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACAAGCGCCGCUCCACCUGCACA---GA .((((((((((((......))))))))).....(((.(((....)))...)))...((((.((((((.(((((((.(((.(((...))).)))..))))))))))))))))).))---). ( -57.30) >DroYak_CAF1 11812 117 + 1 ACUGGCCGCUGCAGAAACGUGCG---GCUGAGCCAAGGCCAAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACAAGCGCCGCUCCACCUGCACACCAGA .((((..((((((......))))---))...(((((......))))).........((((.((((((.(((((((.(((.(((...))).)))..)))))))))))))))))...)))). ( -52.10) >consensus ACUGGCCGCUGCAGAAACGUGCG___GCUGACCCAAGGCCAAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACA_GCGCCGCUCCACCUGCACA___GA ...(((...((((......))))...)))....(((.(((....)))...)))...((((.((((((.((((((..(((.(((...))).)))...))))))))))))))))........ (-37.38 = -38.08 + 0.70)

| Location | 16,733,105 – 16,733,201 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -30.94 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16733105 96 + 22224390 AAUUGGCAAAUUGAAAGCGGAGUGGAGUCGUCGUUCUAAUUGGAAUCCACUUACA-GCGCCGCUCCACCUGCACA---GAAUC--------------------AUUUGGAUGGAUGGCAA .....((.........((((.((((((.((.((((.(((.(((...))).))).)-))).)))))))))))).((---..(((--------------------.....)))...)))).. ( -29.90) >DroSec_CAF1 20852 96 + 1 AAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACA-GCGCCGCUCCACCUGCACA---GAAUC--------------------ACUUGGAUGUGUGGCAA ..(((.((........((((.((((((.(((((((.(((.(((...))).))).)-))))))))))))))))...---....(--------------------((......))))).))) ( -37.60) >DroSim_CAF1 18235 96 + 1 AAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACA-GCGCCGCUCCACCUGCACA---GAAUC--------------------ACUUGGAUGUGUGGCAA ..(((.((........((((.((((((.(((((((.(((.(((...))).))).)-))))))))))))))))...---....(--------------------((......))))).))) ( -37.60) >DroEre_CAF1 12206 97 + 1 AAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACAAGCGCCGCUCCACCUGCACA---GAAUC--------------------AGUUGGAUGUGUGGCAA .....((.........((((.((((((.(((((((.(((.(((...))).)))..)))))))))))))))))(((---..(((--------------------.....))).))).)).. ( -35.10) >DroYak_CAF1 11849 120 + 1 AAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACAAGCGCCGCUCCACCUGCACACCAGAAUCACAGAAUCACAGAAUCGCAGAAUUGAAUGUGUGGCAA .....((.........((((.((((((.(((((((.(((.(((...))).)))..)))))))))))))))))..................((((.(.(((......))).).)))))).. ( -38.40) >consensus AAUUGGCAAAUUGAAAGCGGAGUGGAGUCGGCGUUCUAAUUGGAAUCCACUUACA_GCGCCGCUCCACCUGCACA___GAAUC____________________ACUUGGAUGUGUGGCAA ..(((.((.(((.((.((((.((((((.((((((..(((.(((...))).)))...)))))))))))))))).(....)..........................)).)))...)).))) (-30.94 = -30.98 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:24 2006