
| Sequence ID | X_DroMel_CAF1 |
|---|---|
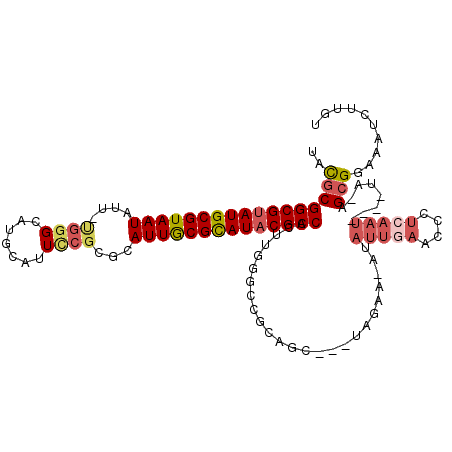
| Location | 16,689,325 – 16,689,430 |
| Length | 105 |
| Max. P | 0.898526 |

| Location | 16,689,325 – 16,689,430 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.75 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16689325 105 + 22224390 UAUGCGGCGUAUGCGUAAUAUU-UGGGCAUGCAUUCCGCGCAUUGCGCAUUCGCCACUUGGGCCGCAGC---UAGAA-AUAUUAAACCCUCAAU----UA-AGCGGAAAUCUUGU ..((((((((((((.((.....-)).)))))).....((((...)))).....((....))))))))..---.....-........((((....----..-)).))......... ( -28.50) >DroSec_CAF1 6009 106 + 1 UAUGCGGCGUAUGCGUAAUAUU-UGGGUAUGCAUUCCGCACAUUGCGCAUACGCCACUUGGUCGGCAGC---UAGAAAAGAUUGAACCCUCAAU----UA-AGCGGAAAUCUUGU ..(((((((((((((((((.(.-((((.......)))).).))))))))))))))..(((((.....))---)))....((((((....)))))----).-.))).......... ( -33.80) >DroSim_CAF1 3174 105 + 1 UAUGCGGCGUAUGCGUAAUAUU-UGGGCAUGCAUUUCGCACAUUGCGCAUACGCCACUUGGGCCGCAGC---UAGAA-AUAUUGAACCCUCAAU----UA-AGCGGAAAUCUUGU .....((((((((((((((...-...((.........))..))))))))))))))((..((.((((...---((...-.))((((....)))).----..-.))))...))..)) ( -32.50) >DroEre_CAF1 5542 102 + 1 GACGCGGCGUAUGCGUAAUAUU-CGGGCUUGCAUUCCGCGCAUUGCGCAUACGCCACUUGGGCC---GC---UAGAA-AUAUUGAACCCUCAAU----UA-GGCGGAAAUCUUGU .....((((((((((((((...-((((.......))))...))))))))))))))((..((.((---((---(....-..(((((....)))))----..-)))))...))..)) ( -38.20) >DroYak_CAF1 5883 106 + 1 AACGCGGCGUAUGCGUAAUAUUUUGGGCAUGCAUUCCGCGCAUUGCGCAUACGCCACUUGGUCCGCAGC---UAGAA-AUAUUGAACCCUCAAU----UA-AGCGGAAAUCUUGU .....((((((((((((((....((((.......))))...))))))))))))))......(((((...---((...-.))((((....)))).----..-.)))))........ ( -34.50) >DroAna_CAF1 10094 103 + 1 CACACGGCGUAUGCGUAAUAUU-CAAG---GCAUUCC-CCUAUUACGUAUACGCCACC------GAAAAAAAAUGAA-ACAAAAAACCUACAAGUAUUUAAGGGGGAAAACUUGU .....((((((((((((((...-..((---(......-)))))))))))))))))...------.............-...........((((((.(((......))).)))))) ( -27.70) >consensus UACGCGGCGUAUGCGUAAUAUU_UGGGCAUGCAUUCCGCGCAUUGCGCAUACGCCACUUGGGCCGCAGC___UAGAA_AUAUUGAACCCUCAAU____UA_AGCGGAAAUCUUGU ..(((((((((((((((((....((((.......))))...)))))))))))))).........................(((((....)))))........))).......... (-24.66 = -25.75 + 1.09)

| Location | 16,689,325 – 16,689,430 |
|---|---|
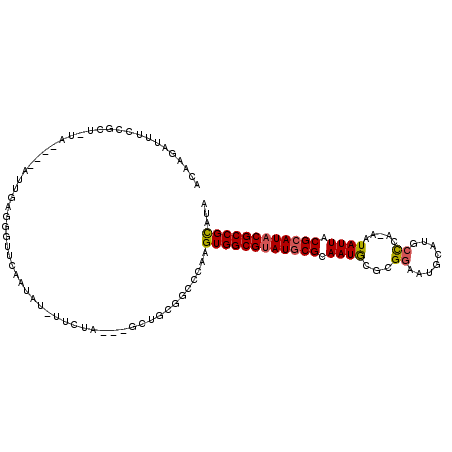
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -21.00 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16689325 105 - 22224390 ACAAGAUUUCCGCU-UA----AUUGAGGGUUUAAUAU-UUCUA---GCUGCGGCCCAAGUGGCGAAUGCGCAAUGCGCGGAAUGCAUGCCCA-AAUAUUACGCAUACGCCGCAUA .........((((.-..----...((((........)-)))..---...)))).....((((((.(((((.((((...((.........)).-..)))).))))).))))))... ( -27.10) >DroSec_CAF1 6009 106 - 1 ACAAGAUUUCCGCU-UA----AUUGAGGGUUCAAUCUUUUCUA---GCUGCCGACCAAGUGGCGUAUGCGCAAUGUGCGGAAUGCAUACCCA-AAUAUUACGCAUACGCCGCAUA ..((((((.((.((-(.----...)))))...)))))).....---............((((((((((((.(((((..((.........)).-.))))).))))))))))))... ( -33.50) >DroSim_CAF1 3174 105 - 1 ACAAGAUUUCCGCU-UA----AUUGAGGGUUCAAUAU-UUCUA---GCUGCGGCCCAAGUGGCGUAUGCGCAAUGUGCGAAAUGCAUGCCCA-AAUAUUACGCAUACGCCGCAUA .........((((.-..----...((((........)-)))..---...)))).....((((((((((((.((((((((.......)))...-.))))).))))))))))))... ( -32.00) >DroEre_CAF1 5542 102 - 1 ACAAGAUUUCCGCC-UA----AUUGAGGGUUCAAUAU-UUCUA---GC---GGCCCAAGUGGCGUAUGCGCAAUGCGCGGAAUGCAAGCCCG-AAUAUUACGCAUACGCCGCGUC .........((((.-..----((((......))))..-.....---))---)).....((((((((((((.((((..(((.........)))-..)))).))))))))))))... ( -34.80) >DroYak_CAF1 5883 106 - 1 ACAAGAUUUCCGCU-UA----AUUGAGGGUUCAAUAU-UUCUA---GCUGCGGACCAAGUGGCGUAUGCGCAAUGCGCGGAAUGCAUGCCCAAAAUAUUACGCAUACGCCGCGUU ........(((((.-..----...((((........)-)))..---...)))))....((((((((((((..(((((.....))))).............))))))))))))... ( -33.16) >DroAna_CAF1 10094 103 - 1 ACAAGUUUUCCCCCUUAAAUACUUGUAGGUUUUUUGU-UUCAUUUUUUUUC------GGUGGCGUAUACGUAAUAGG-GGAAUGC---CUUG-AAUAUUACGCAUACGCCGUGUG ((((((.((........)).))))))...........-.............------...(((((((.(((((((.(-((....)---))..-..))))))).)))))))..... ( -24.70) >consensus ACAAGAUUUCCGCU_UA____AUUGAGGGUUCAAUAU_UUCUA___GCUGCGGCCCAAGUGGCGUAUGCGCAAUGCGCGGAAUGCAUGCCCA_AAUAUUACGCAUACGCCGCAUA ..........................................................((((((((((((.((((...((........)).....)))).))))))))))))... (-21.00 = -21.08 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:53 2006