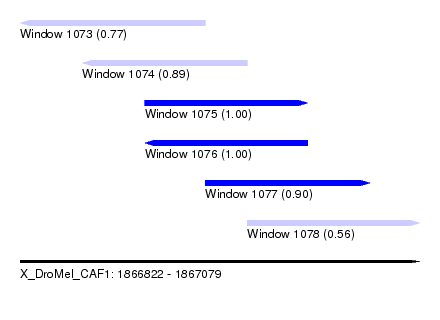
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,866,822 – 1,867,079 |
| Length | 257 |
| Max. P | 0.998051 |

| Location | 1,866,822 – 1,866,941 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -29.34 |
| Energy contribution | -30.15 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1866822 119 - 22224390 UGUGUGUU-UUGUGUUGUGUGUGGCUGCGCCAAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCAGCAAAGAAGAAGCAGCGAUAGGCGAAGUGAGCAAAGAGCAA ..(((.((-((((.((((.(((.((((((((...)))...(((....(((..((.....))..)))......)))...............))))).))).)))).....)))))).))). ( -31.40) >DroSec_CAF1 4063 114 - 1 UGUGUGUU-UUGUGUUGU--GUGGCUGCGCCGAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCAGCAAAG---AAGCAGCGAUAGGCGAAGUGCGCAAAGAGCAA ....((((-((.((((((--(((((...(((...)))..........(((..((.....))..))).....)))))))))))..)---)))))((.....(((.....))).....)).. ( -35.60) >DroEre_CAF1 2653 119 - 1 UGUGUGUUUUUGUGUUGUGUGU-GCUGCGCCAAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCUGCAAAGAAGAAGCAGCGAUAGGCGAAGUGCGCAAAGAGCAA .........((((....((((.-.((.((((...(((..((....))(((..((.....))..))).....))).((((((.........))))))...)))).))..))))....)))) ( -38.90) >DroYak_CAF1 4536 120 - 1 UGUGUGUUUUUGUGUUGUGUGUGGCUGCGCCAAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCAGCAAAGAAGAAGCAGCGAUAGGCAAAGUGCGCAAAGAGCAG (((((((((((((....(((.((((((((((((......))))....)))...))))))))..))))))))(((.(((.((.........)).)))...))).....)))))........ ( -33.40) >consensus UGUGUGUU_UUGUGUUGUGUGUGGCUGCGCCAAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCAGCAAAGAAGAAGCAGCGAUAGGCGAAGUGCGCAAAGAGCAA (((((((((((((.((((...((((((((((((......))))....)))...))))))))).))))))))(((.(((.((.........)).)))...))).....)))))........ (-29.34 = -30.15 + 0.81)

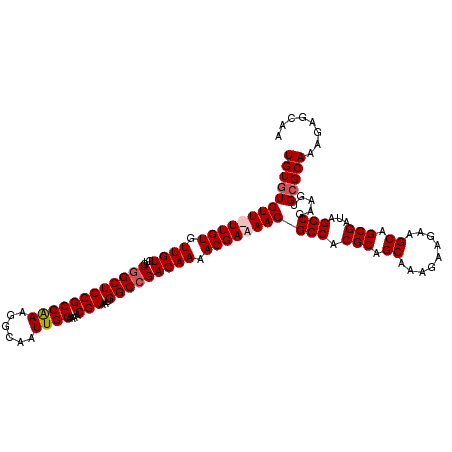
| Location | 1,866,862 – 1,866,968 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -31.41 |
| Energy contribution | -32.22 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1866862 106 - 22224390 ---U----------ACAUUGGCAGCCGCACACACACGCUUUGUGUGUU-UUGUGUUGUGUGUGGCUGCGCCAAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCAG ---(----------((....(((((((((((((.((((..........-..)))))))))))))))))(((...)))...........))).((((((......)).....))))..... ( -36.50) >DroSec_CAF1 4100 104 - 1 ---U----------ACAUUGGCAGCCGCACACACACGCUUUGUGUGUU-UUGUGUUGU--GUGGCUGCGCCGAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCAG ---(----------((....((((((((((((((((((.....)))).-..))).)))--))))))))(((...)))...........))).((((((......)).....))))..... ( -35.20) >DroEre_CAF1 2693 119 - 1 AACCACACAUACACACUUUGGCAGCCGCACACACACGCUUUGUGUGUUUUUGUGUUGUGUGU-GCUGCGCCAAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCUG ...............(((((((.((.((((((((((((.............))).)))))))-)).)))))))))((...(((....(((..((.....))..)))......))).)).. ( -36.92) >DroYak_CAF1 4576 110 - 1 AAAC----------ACAUUGGCUGCCGCACACACACGCUUUGUGUGUUUUUGUGUUGUGUGUGGCUGCGCCAAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCAG ....----------...(((((.((((((((((.((((.............))))))))))))))...))))).(((..((....))(((..((.....))..))).....)))...... ( -34.82) >consensus ___C__________ACAUUGGCAGCCGCACACACACGCUUUGUGUGUU_UUGUGUUGUGUGUGGCUGCGCCAAAGGCAAUUGGAAAACGUAAUGGCCGACAAAACGAAAACGCCACGCAG .................((((((((((((((((.((((.............)))))))))))))))..))))).(((..((....))(((..((.....))..))).....)))...... (-31.41 = -32.22 + 0.81)

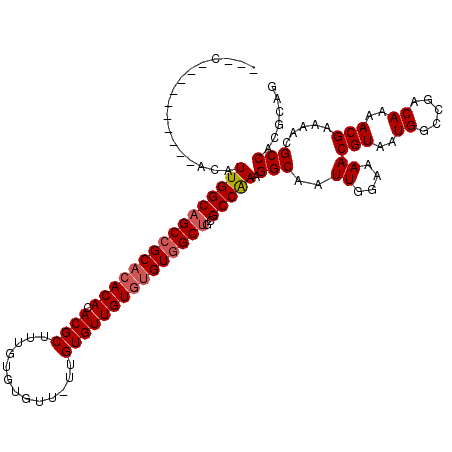
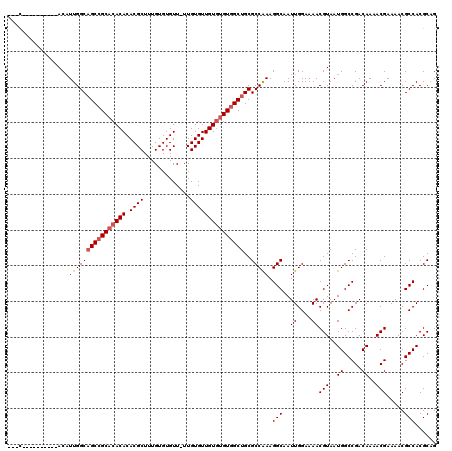
| Location | 1,866,902 – 1,867,007 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -25.91 |
| Energy contribution | -27.54 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1866902 105 + 22224390 AUUGCCUUUGGCGCAGCCACACACAACACAA-AACACACAAAGCGUGUGUGUGCGGCUGCCAAUGU----------A----GAUGUACAUGUUGAUGUGCGCCCGCAAAGAAAAAUCGUU .((((....(((((((((.((((((.(((..-............))))))))).))))))......----------.----...((((((....))))))))).))))............ ( -37.34) >DroSec_CAF1 4140 97 + 1 AUUGCCUUCGGCGCAGCCAC--ACAACACAA-AACACACAAAGCGUGUGUGUGCGGCUGCCAAUGU----------A----GAUGUGCAUGUAGAUG------CGCAAAGAAAAAUCGUU ...(((...)))((((((.(--((.......-.((((((.....))))))))).))))))......----------.----..(((((((....)))------))))............. ( -32.60) >DroEre_CAF1 2733 119 + 1 AUUGCCUUUGGCGCAGC-ACACACAACACAAAAACACACAAAGCGUGUGUGUGCGGCUGCCAAAGUGUGUAUGUGUGGUUGUGUGUGCAUGUAAAUGUACAACCGCAAAGAAAAAUCGUU ...(((((((((((.((-(((((((.(.(.............).)))))))))).)).))))))).)).....((((((((((..(........)..))))))))))............. ( -44.72) >DroYak_CAF1 4616 110 + 1 AUUGCCUUUGGCGCAGCCACACACAACACAAAAACACACAAAGCGUGUGUGUGCGGCAGCCAAUGU----------GUUUGUAUGUGCAUGUAAAUGUGCGCCAGCAAAGAAAAAUCGUU .((((...(((((((..((((.((((((((...(((((((.....)))))))((....))...)))----------).)))).))))(((....))))))))))))))............ ( -36.00) >consensus AUUGCCUUUGGCGCAGCCACACACAACACAA_AACACACAAAGCGUGUGUGUGCGGCUGCCAAUGU__________A____GAUGUGCAUGUAAAUGUGCGCCCGCAAAGAAAAAUCGUU .((((.....(.((((((.((((((.(((...............))))))))).)))))))......................(((((((....)))))))...))))............ (-25.91 = -27.54 + 1.63)

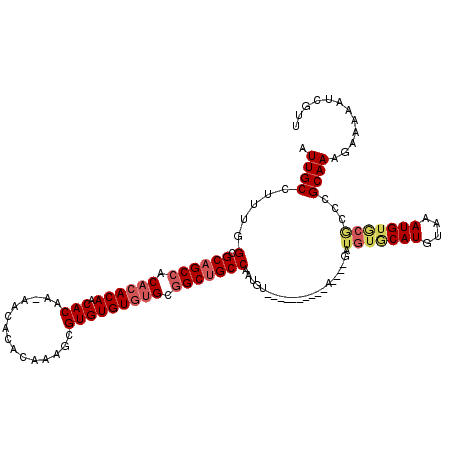
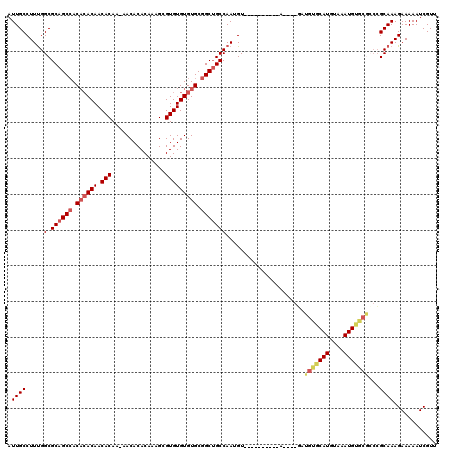
| Location | 1,866,902 – 1,867,007 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -27.41 |
| Energy contribution | -28.22 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1866902 105 - 22224390 AACGAUUUUUCUUUGCGGGCGCACAUCAACAUGUACAUC----U----------ACAUUGGCAGCCGCACACACACGCUUUGUGUGUU-UUGUGUUGUGUGUGGCUGCGCCAAAGGCAAU ............((((.(((.........((((((....----)----------))).))(((((((((((((.((((..........-..))))))))))))))))))))....)))). ( -39.10) >DroSec_CAF1 4140 97 - 1 AACGAUUUUUCUUUGCG------CAUCUACAUGCACAUC----U----------ACAUUGGCAGCCGCACACACACGCUUUGUGUGUU-UUGUGUUGU--GUGGCUGCGCCGAAGGCAAU ..((((.......((.(------(((....)))).))..----.----------..))))((((((((((((((((((.....)))).-..))).)))--))))))))(((...)))... ( -35.60) >DroEre_CAF1 2733 119 - 1 AACGAUUUUUCUUUGCGGUUGUACAUUUACAUGCACACACAACCACACAUACACACUUUGGCAGCCGCACACACACGCUUUGUGUGUUUUUGUGUUGUGUGU-GCUGCGCCAAAGGCAAU ............((((((((((................))))))...........(((((((.((.((((((((((((.............))).)))))))-)).))))))))))))). ( -41.81) >DroYak_CAF1 4616 110 - 1 AACGAUUUUUCUUUGCUGGCGCACAUUUACAUGCACAUACAAAC----------ACAUUGGCUGCCGCACACACACGCUUUGUGUGUUUUUGUGUUGUGUGUGGCUGCGCCAAAGGCAAU ............((((((((((((((....)))(((((((((.(----------(((...((....))(((((((.....)))))))...)))))))))))))..))))))...))))). ( -38.10) >consensus AACGAUUUUUCUUUGCGGGCGCACAUCUACAUGCACAUA____C__________ACAUUGGCAGCCGCACACACACGCUUUGUGUGUU_UUGUGUUGUGUGUGGCUGCGCCAAAGGCAAU .........(((((((.......(((....)))...........................(((((((((((((.((((.............)))))))))))))))))).)))))).... (-27.41 = -28.22 + 0.81)

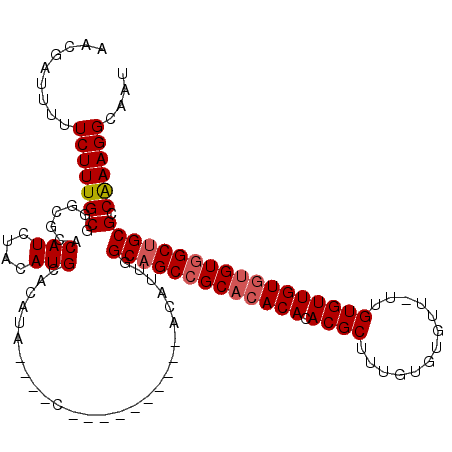
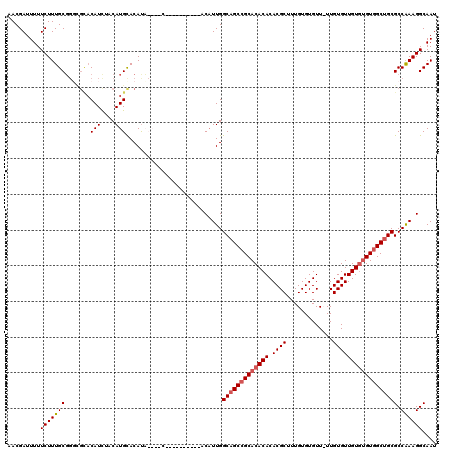
| Location | 1,866,941 – 1,867,047 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -28.21 |
| Energy contribution | -29.15 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1866941 106 + 22224390 AAGCGUGUGUGUGCGGCUGCCAAUGU----------A----GAUGUACAUGUUGAUGUGCGCCCGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUCAAGUGGGCGUCGUGCGAAAGUA ..(((((..((((((.((((....))----------)----).))))))..)..))))((((((((..((((((....))))))...((((....)))).))))))))..(((....))) ( -34.20) >DroSec_CAF1 4177 100 + 1 AAGCGUGUGUGUGCGGCUGCCAAUGU----------A----GAUGUGCAUGUAGAUG------CGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUCAAGUGGGCGUCGUGCGAAAGCA ..((.((..((..((.((((....))----------)----).))..))..))((((------(.((.((((((....))))))...((((....))))..)).)))))))((....)). ( -29.50) >DroEre_CAF1 2772 120 + 1 AAGCGUGUGUGUGCGGCUGCCAAAGUGUGUAUGUGUGGUUGUGUGUGCAUGUAAAUGUACAACCGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUCAAGUGGGCGUCGUGCGAAAGCA ..((.....((..((((.(((....((.(((..((((((((((..(........)..)))))))))).((((((....)))))).........))).))....)))))))..))...)). ( -38.60) >DroYak_CAF1 4656 110 + 1 AAGCGUGUGUGUGCGGCAGCCAAUGU----------GUUUGUAUGUGCAUGUAAAUGUGCGCCAGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUUAAGUGGGCGUCGUGCGAAAGCA ..((.....((..((((.(((.....----------........(..(((....)))..).(((.(..((((((....)))))).).))).............)))))))..))...)). ( -30.60) >consensus AAGCGUGUGUGUGCGGCUGCCAAUGU__________A____GAUGUGCAUGUAAAUGUGCGCCCGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUCAAGUGGGCGUCGUGCGAAAGCA ..((.....((..((((.(((......................(((((((....))))))).((((..((((((....))))))..)))).............)))))))..))...)). (-28.21 = -29.15 + 0.94)

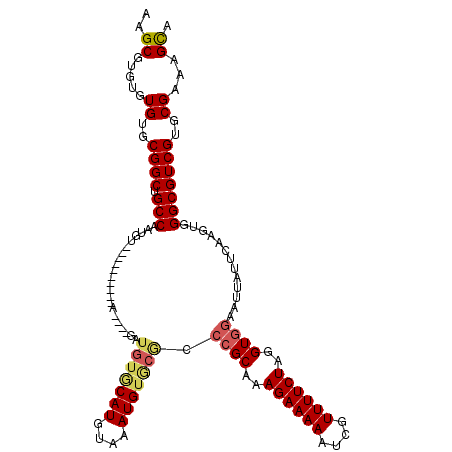
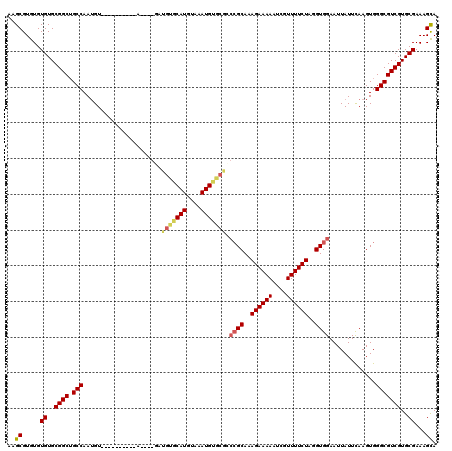
| Location | 1,866,968 – 1,867,079 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.27 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -21.29 |
| Energy contribution | -22.60 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1866968 111 + 22224390 -GAUGUACAUGUUGAUGUGCGCCCGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUCAAGUGGGCGUCGUGCGAAAGUAUUUCCGUGGCG-AAAACUAGUUUAAGAAUCGCG------- -(((((((((....))))))((((((..((((((....))))))...((((....)))).)))))))))((((....)))).......(((-(...((......))..)))).------- ( -31.30) >DroSec_CAF1 4204 106 + 1 -GAUGUGCAUGUAGAUG------CGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUCAAGUGGGCGUCGUGCGAAAGCAUUUCCGUGGCGCAAAACUAGUUUAGGAAUCGCA------- -..(((((((....)))------)))).((((((....))))))..((((..(((..(.(((..(((((((((....))))......)))))...))).)..)))...)))).------- ( -30.90) >DroEre_CAF1 2812 120 + 1 GUGUGUGCAUGUAAAUGUACAACCGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUCAAGUGGGCGUCGUGCGAAAGCAUUUCCCUGGCCCAAAACUAGUUUAAGAAUGGCGAGUGAUA ...(((((((....))))))).((((..((((((....))))))..)))).(((((((...((((((..((((....))))..)....)))))......(((.......)))))))))). ( -36.90) >DroYak_CAF1 4686 120 + 1 GUAUGUGCAUGUAAAUGUGCGCCAGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUUAAGUGGGCGUCGUGCGAAAGCAUUUCCCUGGCCCAAAACUAGUUUAAGAAUGACGGUAAUGA ((..((((((......))))))..))........((((((((((.....((((((.((...((((((..((((....))))..)....))))).)).)))))).)))).))))))..... ( -30.40) >consensus _GAUGUGCAUGUAAAUGUGCGCCCGCAAAGAAAAAUCGUUUUCUAGGUGGAAUUAUUCAAGUGGGCGUCGUGCGAAAGCAUUUCCCUGGCCCAAAACUAGUUUAAGAAUCGCG_______ ...(((((((....))))))).((((..((((((....))))))..))))..(((..(.(((..(((((((((....))))......)))))...))).)..)))............... (-21.29 = -22.60 + 1.31)

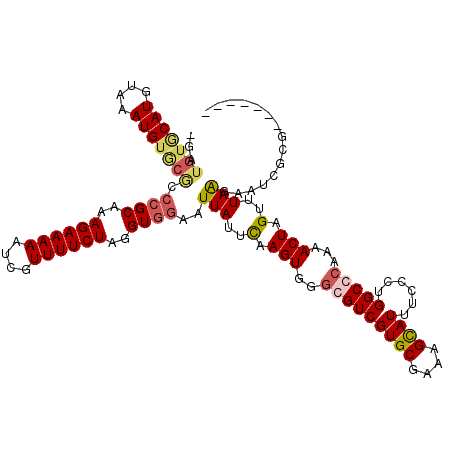
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:44 2006