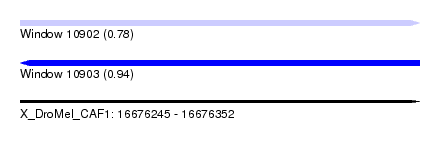
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,676,245 – 16,676,352 |
| Length | 107 |
| Max. P | 0.941060 |

| Location | 16,676,245 – 16,676,352 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16676245 107 + 22224390 UAUGAUUUCUGUU--AACGG---UCUCACCUGCAUGAAUAUAAACUCCCACUUUCUGGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCGGCCAACUCCA ..........(((--...((---((....(((((.(((.((...((((........))))...)).))))))))...))))........((((........))))))).... ( -26.30) >DroVir_CAF1 7377 109 + 1 AACAU---UGAAUUAGCCAUUUAACUCACCUGCAUGAAUAUGAACUCCCAUUUGCGUGAGAACAUCUUCUGCAGCUUGGCCAGGUUCUCGGCUUCGUAGUCCGCCAACUCCA .....---.(((((.((((..........(((((.(((.(((..(((.(......).)))..))).))))))))..))))..)))))..(((..........)))....... ( -21.60) >DroPse_CAF1 21575 104 + 1 CAGUUCC--UAUU-CGAU-----ACUUACCUGCAUGAAGAUGAACUCCCAUUUCCGAGAGAACAUCUUCUGCAACUUGGCCAGGUUCUCGGCUUCGUAGUCGGCCAAUUCCA .......--....-....-----.......((((.(((((((..(((.(......).)))..)))))))))))..((((((..(....)((((....))))))))))..... ( -27.60) >DroEre_CAF1 1256 103 + 1 UACAUUUUAUGUU--G-------GCCUACCUGCAUGAAUAUAAACUCCCACUUCCUGGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCAUAGUCGGCCAACUCGA ..........(((--(-------(((.(((((((.(((.((...((((........))))...)).))))))))...((((........)))).....)).))))))).... ( -32.70) >DroMoj_CAF1 10443 109 + 1 AAUAUA-UCAAAU--GAUAUCCCACACACCUGCAUGAAUAUGAACUCCCAUUUGCGCGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCCGCCAGCUCCA ...(((-((....--))))).........(((((.(((.(((..(((.(......).)))..))).)))))))).(((((..((((..((....)).)))).)))))..... ( -21.60) >DroPer_CAF1 19801 104 + 1 CAGUUCC--UAUU-CGAU-----ACUUACCUGCAUGAAGAUGAACUCCCAUUUCCGAGAGAACAUCUUCUGCAACUUGGCCAGGUUCUCGGCUUAGUAGUCGGCCAAUUCCA .......--...(-((((-----.......((((.(((((((..(((.(......).)))..)))))))))))(((.((((........)))).))).)))))......... ( -27.70) >consensus AACAUC___UAUU__GAU_____ACUCACCUGCAUGAAUAUGAACUCCCAUUUCCGAGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCGGCCAACUCCA .............................(((((.(((.(((..(((..........)))..))).)))))))).((((((.(.....)(((......)))))))))..... (-16.44 = -17.30 + 0.86)

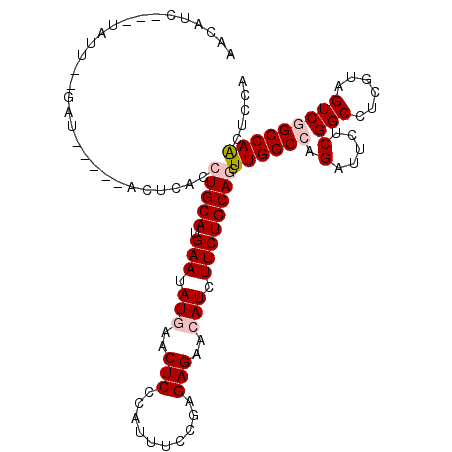
| Location | 16,676,245 – 16,676,352 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -22.41 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16676245 107 - 22224390 UGGAGUUGGCCGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCCAGAAAGUGGGAGUUUAUAUUCAUGCAGGUGAGA---CCGUU--AACAGAAAUCAUA .((..((((((........))))))...(((.(((....(((((((.(((..((((........))))..))).))).))))))).)))---))...--............. ( -31.90) >DroVir_CAF1 7377 109 - 1 UGGAGUUGGCGGACUACGAAGCCGAGAACCUGGCCAAGCUGCAGAAGAUGUUCUCACGCAAAUGGGAGUUCAUAUUCAUGCAGGUGAGUUAAAUGGCUAAUUCA---AUGUU ..((((((((.((((((...((((......))))....((((((((.(((..(((.((....)).)))..))).))).))))))).)))).....)))))))).---..... ( -31.30) >DroPse_CAF1 21575 104 - 1 UGGAAUUGGCCGACUACGAAGCCGAGAACCUGGCCAAGUUGCAGAAGAUGUUCUCUCGGAAAUGGGAGUUCAUCUUCAUGCAGGUAAGU-----AUCG-AAUA--GGAACUG .....(((((((.((.((....))))....))))))).((((((((((((..((((((....))))))..))))))).)))))...(((-----.((.-....--)).))). ( -34.50) >DroEre_CAF1 1256 103 - 1 UCGAGUUGGCCGACUAUGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCCAGGAAGUGGGAGUUUAUAUUCAUGCAGGUAGGC-------C--AACAUAAAAUGUA ....(((((((.((.....(((((......)))))...((((((((.(((..((((........))))..))).))).))))))).)))-------)--))).......... ( -40.00) >DroMoj_CAF1 10443 109 - 1 UGGAGCUGGCGGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCGCGCAAAUGGGAGUUCAUAUUCAUGCAGGUGUGUGGGAUAUC--AUUUGA-UAUAUU ((.((((..((.....)).(((((......))))).)))).))...(((((((.((((((..((.((((....))))...))..)))))))))))))--......-...... ( -35.60) >DroPer_CAF1 19801 104 - 1 UGGAAUUGGCCGACUACUAAGCCGAGAACCUGGCCAAGUUGCAGAAGAUGUUCUCUCGGAAAUGGGAGUUCAUCUUCAUGCAGGUAAGU-----AUCG-AAUA--GGAACUG .....(((((((....((......))....))))))).((((((((((((..((((((....))))))..))))))).)))))...(((-----.((.-....--)).))). ( -33.80) >consensus UGGAGUUGGCCGACUACGAAGCCGAGAACCUGGCCAAGCUGCAGAAGAUGUUCUCCCGGAAAUGGGAGUUCAUAUUCAUGCAGGUAAGU_____AUC__AAUA___AAUAUA .....(((((((.((.((....))))....))))))).((((((((.(((..((((((....))))))..))).))).)))))............................. (-22.41 = -23.05 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:48 2006