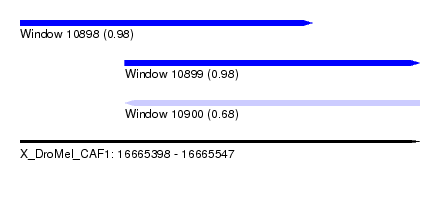
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,665,398 – 16,665,547 |
| Length | 149 |
| Max. P | 0.981874 |

| Location | 16,665,398 – 16,665,507 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16665398 109 + 22224390 AAUUUGGAAUAUAAGUAGUUCCACACACU-UCCAUUCACAAAUUGCAUCAGCAUCGGGUCGAAAAUCG-AUUCGAGUCAUUGACCAGAUGUUGAAAAGCACAUUAUAUAUC ((((((((((..((((.((.....)))))-)..)))).))))))((.((((((((.((((((...(((-...)))....)))))).))))))))...))............ ( -28.90) >DroSec_CAF1 4339 85 + 1 AAUU--------------------------UCCAUUCACAAAUUGCAUCAGCAUCGGGUCGAAAAUCGGGUUCGAGUCAUUGACCAGAUGUUGAAAAGCACAUUAUAUAUC ....--------------------------.............(((.((((((((.((((((...(((....)))....)))))).))))))))...)))........... ( -23.10) >DroEre_CAF1 5611 104 + 1 AAUUUUGAAUACAAUU-----CGCAUGCU-UCCAUUCACAAAUUGCACCAGCUUUGGGUCGAGGAUCG-GAUCGAGUCAUUGACCCGAUGUUGAAGAGCAAAUUAUAUUUC ......(((((.((((-----.((.(((.-..............))).((((.(((((((((.(((((-...))).)).))))))))).))))....)).)))).))))). ( -25.86) >DroYak_CAF1 4123 105 + 1 AAUUUUGAAUACAAUU-----CGCAUGCUUUCCAUUCACAAAUUGCAUCAGCAACGGGUCGAAAAUCG-GAUCGAGUCAUUGACCAGAUGUUGAAAAGCACAUCACAUUUC ................-----.((.(((................)))((((((.(.((((((...(((-...)))....)))))).).))))))...))............ ( -19.09) >consensus AAUUUUGAAUACAAUU_____CGCAUGCU_UCCAUUCACAAAUUGCAUCAGCAUCGGGUCGAAAAUCG_GAUCGAGUCAUUGACCAGAUGUUGAAAAGCACAUUAUAUAUC ...........................................(((.((((((((.((((((...(((....)))....)))))).))))))))...)))........... (-19.21 = -19.78 + 0.56)


| Location | 16,665,437 – 16,665,547 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -20.69 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16665437 110 + 22224390 AAUUGCAUCAGCAUCGGGUCGAAAAUCG-AUUCGAGUCAUUGACCAGAUGUUGAAAAGCACAUUAUAUAUCCAGAGGUUAAGUGUGUGUGUGUGUGUGUGAUAUGGUCAUA .((..(.((((((((.((((((...(((-...)))....)))))).))))))))...((((((.(((((..((.........))..))))))))))))..))......... ( -31.90) >DroSec_CAF1 4353 101 + 1 AAUUGCAUCAGCAUCGGGUCGAAAAUCGGGUUCGAGUCAUUGACCAGAUGUUGAAAAGCACAUUAUAUAUCCAGAGGUUAAGUAUAUAUAUA----UACAAU------AUA ...(((.((((((((.((((((...(((....)))....)))))).))))))))...))).....................(((((....))----)))...------... ( -25.60) >DroEre_CAF1 5645 102 + 1 AAUUGCACCAGCUUUGGGUCGAGGAUCG-GAUCGAGUCAUUGACCCGAUGUUGAAGAGCAAAUUAUAUUUCCAAACGUUAAGUAUGUGUG---GUAUGA--UAUAUU---A ..((((..((((.(((((((((.(((((-...))).)).))))))))).))))....))))..((((((.(((.((((.....)))).))---)...))--))))..---. ( -30.70) >DroYak_CAF1 4158 107 + 1 AAUUGCAUCAGCAACGGGUCGAAAAUCG-GAUCGAGUCAUUGACCAGAUGUUGAAAAGCACAUCACAUUUCCAGACGUUAAGUAUGUGUG---AUAUGGGAUAUACUUUAG ...(((.((((((.(.((((((...(((-...)))....)))))).).))))))...))).(((.(((...((.((((.....)))).))---..))).)))......... ( -24.40) >consensus AAUUGCAUCAGCAUCGGGUCGAAAAUCG_GAUCGAGUCAUUGACCAGAUGUUGAAAAGCACAUUAUAUAUCCAGACGUUAAGUAUGUGUG___GUAUGAGAUAUA_U_AUA ...(((.((((((((.((((((...(((....)))....)))))).))))))))...)))...(((((((.(.........).)))))))..................... (-20.69 = -20.62 + -0.06)


| Location | 16,665,437 – 16,665,547 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -14.45 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16665437 110 - 22224390 UAUGACCAUAUCACACACACACACACACACUUAACCUCUGGAUAUAUAAUGUGCUUUUCAACAUCUGGUCAAUGACUCGAAU-CGAUUUUCGACCCGAUGCUGAUGCAAUU .......(((((.((.......................)))))))......(((...(((.((((.((((...((.(((...-))).))..)))).)))).))).)))... ( -17.70) >DroSec_CAF1 4353 101 - 1 UAU------AUUGUA----UAUAUAUAUACUUAACCUCUGGAUAUAUAAUGUGCUUUUCAACAUCUGGUCAAUGACUCGAACCCGAUUUUCGACCCGAUGCUGAUGCAAUU .((------((((((----(((...................)))))))))))((...(((.((((.((((...((.(((....))).))..)))).)))).))).)).... ( -23.01) >DroEre_CAF1 5645 102 - 1 U---AAUAUA--UCAUAC---CACACAUACUUAACGUUUGGAAAUAUAAUUUGCUCUUCAACAUCGGGUCAAUGACUCGAUC-CGAUCCUCGACCCAAAGCUGGUGCAAUU .---..((((--(....(---((.((.........)).)))..)))))..((((........((((((((........))))-)))).......(((....))).)))).. ( -17.50) >DroYak_CAF1 4158 107 - 1 CUAAAGUAUAUCCCAUAU---CACACAUACUUAACGUCUGGAAAUGUGAUGUGCUUUUCAACAUCUGGUCAAUGACUCGAUC-CGAUUUUCGACCCGUUGCUGAUGCAAUU ..((((((((((.(((.(---(.((.((.......)).)))).))).))))))))))....((((.((.(((((..((((..-......))))..)))))))))))..... ( -25.00) >consensus UAU_A_UAUAUCACAUAC___CACACAUACUUAACCUCUGGAAAUAUAAUGUGCUUUUCAACAUCUGGUCAAUGACUCGAAC_CGAUUUUCGACCCGAUGCUGAUGCAAUU ...................................................(((...(((.((((.((((...((.(((....))).))..)))).)))).))).)))... (-14.45 = -15.08 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:45 2006