
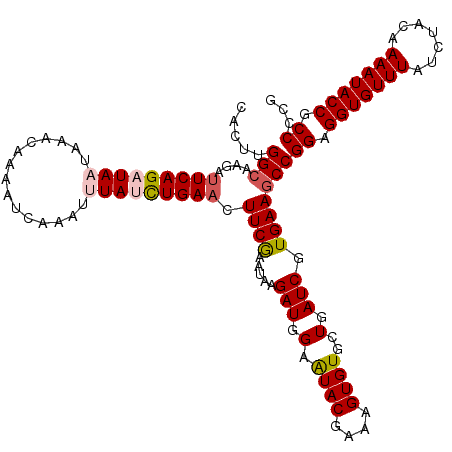
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,634,483 – 16,634,637 |
| Length | 154 |
| Max. P | 0.998425 |

| Location | 16,634,483 – 16,634,601 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -25.40 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16634483 118 - 22224390 CACUUGGAAAGAUUCAGAUAAUAAACAAAAUCAAAUUUAUCUGAACUUCGAAUAAGAUGGAAUACGAAAGUGUGCUGAUCGUGAAGCCGGAGGUGUUUAUCUACAAAAUACCACCCCG .....((.....(((((((((...............)))))))))(((((.....(((.(.((((....))))..).))).)))))))((.(((((((.......))))))).))... ( -28.86) >DroSec_CAF1 1954 118 - 1 CACUUGGCAAGAUUCAGAUAAUAAACAAAAUCAAAUUUAUCUGAACUUCGAAUAAGAUGGAAUACGAAAGUGUGCUGAUCGUGAAGCCGGAGGUGUUUAUCUACAAAAUACCGCCCCG .....((((((.(((((((((...............)))))))))))).......(((.(.((((....))))..).))).....)))((.(((((((.......))))))).))... ( -31.06) >DroSim_CAF1 1960 118 - 1 CACUUGGCAAGACUCAGAUAAUAAACAAAAUCAAAUUUAUCUGAACUUCGAAUAAGAUGGAAUACGAAAGUGUGCUGAUCGUGAAGCCGGAGGUGUUUAUCUACAAAAUACCGCCCCG .....(((.....((((((((...............))))))))..((((.....(((.(.((((....))))..).))).)))))))((.(((((((.......))))))).))... ( -29.96) >DroEre_CAF1 2163 118 - 1 CACUUGGCAAAAUUCAGAUAAUAAACAAAGUCAAAAAUAGUUGAAUUUCAAAUAAGAUGGAAUACGAAAGUGUGCUGAUCGUGAAGCCGGAGGUGUUUAUCUACAAAAUACCGCCCCG .....(((......................((((......))))..((((.....(((.(.((((....))))..).))).)))))))((.(((((((.......))))))).))... ( -24.70) >DroYak_CAF1 1982 118 - 1 CACUUGGCAAAAUUCAGAUAAUAAACAAAAUCAAAAAUAGUUGAAUUUCGAAUAAGAUGGAGUACGAAAGUGUGCUGAUCGUGAAGCCGGAGGUGUUUAUCUACAAAAUACCGCCCCG .....(((......................((((......))))..((((.....(((((..(((....)))..)).))).)))))))((.(((((((.......))))))).))... ( -23.60) >consensus CACUUGGCAAGAUUCAGAUAAUAAACAAAAUCAAAUUUAUCUGAACUUCGAAUAAGAUGGAAUACGAAAGUGUGCUGAUCGUGAAGCCGGAGGUGUUUAUCUACAAAAUACCGCCCCG .....(((....(((((((((...............))))))))).((((.....(((.(.((((....))))..).))).)))))))((.(((((((.......))))))).))... (-25.40 = -26.04 + 0.64)

| Location | 16,634,523 – 16,634,637 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16634523 114 - 22224390 UCGCCCGCCGGUCACACUGGCUAAAGCUUAAA----AAAACACUUGGAAAGAUUCAGAUAAUAAACAAAAUCAAAUUUAUCUGAACUUCGAAUAAGAUGGAAUACGAAAGUGUGCUGA ......(((((.....)))))...(((.....----....((((((..(((.(((((((((...............))))))))))))....)))).))..((((....))))))).. ( -27.06) >DroSec_CAF1 1994 114 - 1 UCGCCCGCCGGUCACACUGGCUAAAGCUUAAA----AAAACACUUGGCAAGAUUCAGAUAAUAAACAAAAUCAAAUUUAUCUGAACUUCGAAUAAGAUGGAAUACGAAAGUGUGCUGA ......(((((.....)))))...(((.....----....((((((..(((.(((((((((...............))))))))))))....)))).))..((((....))))))).. ( -27.06) >DroSim_CAF1 2000 115 - 1 UCGCCCGCCGGUCACACUGGCUAAAGCUUAAAA---AAAACACUUGGCAAGACUCAGAUAAUAAACAAAAUCAAAUUUAUCUGAACUUCGAAUAAGAUGGAAUACGAAAGUGUGCUGA ......(((((.....)))))...(((......---....((((((....((.((((((((...............))))))))...))...)))).))..((((....))))))).. ( -25.16) >DroEre_CAF1 2203 118 - 1 UCGCCCGCCGGUCACACUGGCUAAAGCUUAAAAAAAAAAACACUUGGCAAAAUUCAGAUAAUAAACAAAGUCAAAAAUAGUUGAAUUUCAAAUAAGAUGGAAUACGAAAGUGUGCUGA ......(((((.....)))))...(((.............((((((...((((((((.......................))))))))....)))).))..((((....))))))).. ( -20.40) >DroYak_CAF1 2022 115 - 1 UCGCUCGCCGGUCACACUGGCUAAAGCUUAAAA---AAAACACUUGGCAAAAUUCAGAUAAUAAACAAAAUCAAAAAUAGUUGAAUUUCGAAUAAGAUGGAGUACGAAAGUGUGCUGA ..(((((((((.....)))))............---....((((((...((((((((.......................))))))))....)))).))))))((....))....... ( -19.90) >consensus UCGCCCGCCGGUCACACUGGCUAAAGCUUAAAA___AAAACACUUGGCAAGAUUCAGAUAAUAAACAAAAUCAAAUUUAUCUGAACUUCGAAUAAGAUGGAAUACGAAAGUGUGCUGA ......(((((.....)))))...(((.............((((((..(((.(((((((((...............))))))))))))....)))).))..((((....))))))).. (-22.06 = -22.18 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:33 2006