| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,619,711 – 16,619,831 |
| Length | 120 |
| Max. P | 0.551746 |

| Location | 16,619,711 – 16,619,831 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
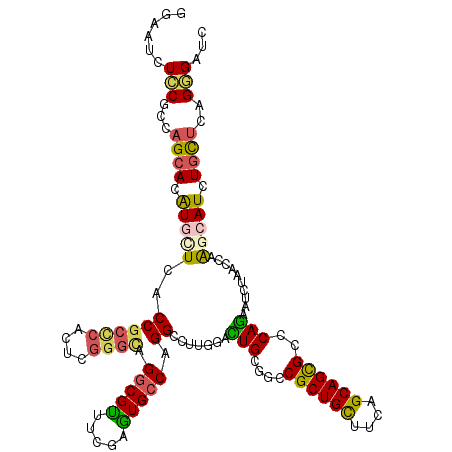
| Mean single sequence MFE | -46.53 |
| Consensus MFE | -26.59 |
| Energy contribution | -26.18 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
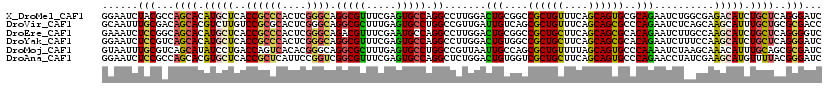
>X_DroMel_CAF1 16619711 120 - 22224390 GGAAUCUACGCCAGCACAUGCUCACCGCCCACUCGGGCAGGCGUUUCGAGUGCCAGGCCUUGGACUGCGGCCGCUGUUUCAGCAGUGCGCAGAAUCUGGCGAGACAUCUGCUCAGGGAUC ...((((((((((((....)))....((((....)))).)))))...(((((((((((....).(((((..((((((....)))))))))))..))))))((....)).))))..)))). ( -46.40) >DroVir_CAF1 1889 120 - 1 GCAAUUUGCGACAGCACGUCUUGUCCGCGCACUCGGGCAGGCGCUUUGAGUGCCUGGCCGUUGAUUGUCAGCGCUGUUUCAGCAGCGCCCAGAAUCUCAGCAAGCAUUUGCUGCGCGACC .....(((((.(((((.(((((((.........(((.(((((((.....))))))).)))..((((.(..(((((((....)))))))..).))))...))))).)).)))))))))).. ( -53.60) >DroEre_CAF1 1869 120 - 1 GAAAUCUCCGGCAGCACAUGCUCACCGCCCACUCGGGCAGACGUUUCGAAUGCCAGGCCUUGGACUGCGGCCGCUGCUUCAGCAGCGCACAGAAUCUUGCCAAGCAUCUGCUCAGGGGUC .....((((.(.((((.(((((....((((....))))....(.(((...(((..((((.........))))(((((....))))))))..))).)......))))).))))).)))).. ( -43.50) >DroYak_CAF1 1670 120 - 1 GGAAUCUCCGUCAGCACAUGCUCACCGCCCACUCGGGCAGGCGUUUCGAGUGCCAGGCCUUGGACUGUGGCCGCUGCUUCAGCAGCGCACAGAAUCUUUCCAAGCAUCUGCUCAGGGAUC ......(((.(.((((.(((((....((((....)))).((...(((..((((..((((.........))))(((((....))))))))).))).....)).))))).)))).).))).. ( -48.00) >DroMoj_CAF1 1991 120 - 1 GUAAUUUGCGUCAGCAUAUCCUGACCAGUCACACGGGCAGGCGCUUUGAGUGCCUGGCCGUUAAUUGCCAGCGCUGUUUUAGCAGUGCCCAAAAUCUAAGCAAACAUUUGCAGCGCGAUC ((...(((.(((((......))))))))..))((((.(((((((.....))))))).))))..((((((.(((((((....)))))))...........((((....)))).).))))). ( -44.90) >DroAna_CAF1 1923 120 - 1 GGAAUCUCCGCCAGCACGUGCUCACCGCUCAUUCCGGUCGGCGUUUCGAGUGCCAGGCUCUGGACUGUGGUCGCUGCUUCAGCAGUGCCCAGAACCUAUCGAAGCAUGUUUUACGGGAUC ...(((.(((..((((...(((.((((.......)))).)))(((((((...((((...)))).(((.((.((((((....)))))))))))......))))))).))))...)))))). ( -42.80) >consensus GGAAUCUCCGCCAGCACAUGCUCACCGCCCACUCGGGCAGGCGUUUCGAGUGCCAGGCCUUGGACUGCGGCCGCUGCUUCAGCAGCGCCCAGAAUCUAACCAAGCAUCUGCUCAGGGAUC ......(((...((((.(((((..((((((....)))).(((((.....))))).)).......(((....((((((....))))))..)))..........))))).))))..)))... (-26.59 = -26.18 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:29 2006