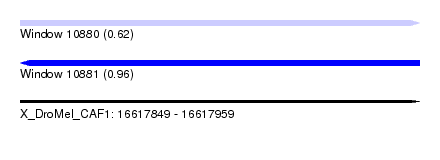
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,617,849 – 16,617,959 |
| Length | 110 |
| Max. P | 0.958964 |

| Location | 16,617,849 – 16,617,959 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -28.77 |
| Energy contribution | -29.15 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16617849 110 + 22224390 CUCGCAGAUCAGCCGCUUGUGCGGUACGUUUUAGGUCCACGGCAUACCUGCUGCCAUUGGUCACACUGGCAGAUUACGCGAUUAUUUU-UUUUACGGAAAGUCUGCGUUUU ..(((((((..(((((....))))).(((..(((((.........)))))((((((.((....)).)))))).....)))........-...........))))))).... ( -34.00) >DroSim_CAF1 28032 110 + 1 CUCGCAGAUCAGCCGUUUGUGCGGUAUGUUUUAGGUGCACGGCAUACCAGCUGCCACUGGUCACACUGGCAGAUUACGCGAUUAUUUU-UUUUACGGAAAGUCUGCGUUUU ..(((((((...(((((((((.(((((((..(......)..)))))))..((((((.((....)).))))))....))))).......-....))))...))))))).... ( -37.90) >DroEre_CAF1 29703 107 + 1 ---GGAGAUCAGCCGCUUGUGCGGUACGUUUUAGGUACACCACGCACUUGCUGCCCUUGGUCACACUGGCAGAUUACGCGAUUAUUUU-UUUUACGGAAAGUCUGCGUUUU ---...(((((((.((..(((((((((.......))))....)))))..)).))...)))))......(((((((.((..(.......-..)..))...)))))))..... ( -29.70) >DroYak_CAF1 31770 111 + 1 CUCGCAGAUCAGCCGUUUCUGCGGUACGUUUUAGGCACACGGCAUGCUUGCUGCCACUGGUCACACUGGCAGAUUACGCGAUUAUUUUUUUUUACGGAAAGUCUGCGUUUU ..(((((((...((((.((.(((........((((((.......))))))((((((.((....)).))))))....)))))............))))...))))))).... ( -34.12) >consensus CUCGCAGAUCAGCCGCUUGUGCGGUACGUUUUAGGUACACGGCAUACCUGCUGCCACUGGUCACACUGGCAGAUUACGCGAUUAUUUU_UUUUACGGAAAGUCUGCGUUUU ..(((((((..(((((....))))).(((..((((((.......))))))((((((.((....)).)))))).....)))....................))))))).... (-28.77 = -29.15 + 0.38)


| Location | 16,617,849 – 16,617,959 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.44 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16617849 110 - 22224390 AAAACGCAGACUUUCCGUAAAA-AAAAUAAUCGCGUAAUCUGCCAGUGUGACCAAUGGCAGCAGGUAUGCCGUGGACCUAAAACGUACCGCACAAGCGGCUGAUCUGCGAG ....((((((.(..((((....-.........(((((..((((((.((....)).))))))....))))).((((((.......)).))))....))))..).)))))).. ( -35.10) >DroSim_CAF1 28032 110 - 1 AAAACGCAGACUUUCCGUAAAA-AAAAUAAUCGCGUAAUCUGCCAGUGUGACCAGUGGCAGCUGGUAUGCCGUGCACCUAAAACAUACCGCACAAACGGCUGAUCUGCGAG ....((((((.(..((((....-.........(((((..((((((.((....)).))))))....))))).((((..............))))..))))..).)))))).. ( -36.24) >DroEre_CAF1 29703 107 - 1 AAAACGCAGACUUUCCGUAAAA-AAAAUAAUCGCGUAAUCUGCCAGUGUGACCAAGGGCAGCAAGUGCGUGGUGUACCUAAAACGUACCGCACAAGCGGCUGAUCUCC--- ......(((......(((....-......((((((((..(((((..((....))..)))))....)))))))).........)))..((((....)))))))......--- ( -28.31) >DroYak_CAF1 31770 111 - 1 AAAACGCAGACUUUCCGUAAAAAAAAAUAAUCGCGUAAUCUGCCAGUGUGACCAGUGGCAGCAAGCAUGCCGUGUGCCUAAAACGUACCGCAGAAACGGCUGAUCUGCGAG ....((((((.(..((((..............((.....((((((.((....)).))))))...)).(((.(.((((.......))))))))...))))..).)))))).. ( -34.00) >consensus AAAACGCAGACUUUCCGUAAAA_AAAAUAAUCGCGUAAUCUGCCAGUGUGACCAAUGGCAGCAAGUAUGCCGUGUACCUAAAACGUACCGCACAAACGGCUGAUCUGCGAG ....((((((.(..((((..............(((((..((((((.((....)).))))))....))))).((((..............))))..))))..).)))))).. (-27.88 = -28.44 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:28 2006