
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,614,962 – 16,615,158 |
| Length | 196 |
| Max. P | 0.955892 |

| Location | 16,614,962 – 16,615,067 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.98 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16614962 105 - 22224390 GUUUGACGGCCCAUUACGUCACCAUCACCAUCAUCGCGAUGCUGAGCGUAGAACGCGCCAAAAUCUAAGCAUGCAGCAAAAAGAAAAAA---AUCCAAAAGCAAACAU (((((.((((..............(((.((((.....)))).)))(((.....)))))).........((.....))............---........)))))).. ( -20.60) >DroSim_CAF1 25183 108 - 1 GUUUGACGGCCCAUUACGUCACCAUCACCAUCAUCGCGAUGCUGAGCGUAGAACGCGCCAAAAUCUAAGCAUGCAGCAAAAAAAAAAAAAUAAUCCAAAAGCAAACAU (((((.((((..............(((.((((.....)))).)))(((.....)))))).........((.....)).......................)))))).. ( -20.60) >DroEre_CAF1 27169 100 - 1 GUUUGGCGGC-CAUUACGUCACC------AUCGUCGCGAUGCUGAGCGUAGAACACGCCAAAAUCUAAGCAUGCAGCGAAAAUAAAGAG-AAACCCAAGAUCAAACAA .(((((((..-..((((((((.(------((((...))))).))).)))))....)))))))......((.....))............-.................. ( -23.40) >DroYak_CAF1 28854 97 - 1 GUUUGACGGCCCAUUACGUCACC------AGCAGCGCGAUGCUGAGCGUAGAACGCGCCAAAAUCUAAGCAUGCAGCUAAAAAAA-----AACACCAAAAGCAAACAU (((((.(((((..((((((...(------((((......))))).))))))...).)))........(((.....))).......-----..........)))))).. ( -22.80) >consensus GUUUGACGGCCCAUUACGUCACC______AUCAUCGCGAUGCUGAGCGUAGAACGCGCCAAAAUCUAAGCAUGCAGCAAAAAAAAAAAA_AAAUCCAAAAGCAAACAU ((((((((........))))...............((.(((((..((((.....)))).........)))))))............................)))).. (-17.45 = -17.07 + -0.38)


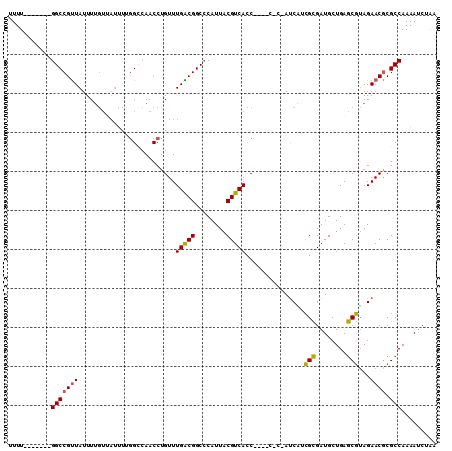
| Location | 16,614,999 – 16,615,100 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16614999 101 + 22224390 UUAGAUUUUGGCGCGUUCUACGCUCAGCAUCGCGAUGAUGGUGA--UGGUGACGUAAUGGGCCGUCAAACAGGUUGGCCAAAAUAACAAAAUAACGGCC-------AAAA .....(((((((.((((.((((.(((.((((((.......))))--)).)))))))...(((((.(......).))))).............)))))))-------)))) ( -34.90) >DroSim_CAF1 25223 101 + 1 UUAGAUUUUGGCGCGUUCUACGCUCAGCAUCGCGAUGAUGGUGA--UGGUGACGUAAUGGGCCGUCAAACAGGUUGGCCGAAAUAACAAAAUAACGGCC-------AAAA ......(((((((.((((((((.(((.((((((.......))))--)).)))))))..)))))))))))....(((((((..............)))))-------)).. ( -36.34) >DroEre_CAF1 27208 94 + 1 UUAGAUUUUGGCGUGUUCUACGCUCAGCAUCGCGACGAU--------GGUGACGUAAUG-GCCGCCAAACAGGUUGCCCAAAAUUACAAAAUAGCGGCC-------AAAA ......(((((((.(((.((((.(((.(((((...))))--------).)))))))..)-)))))))))..((((((................))))))-------.... ( -31.09) >DroYak_CAF1 28889 95 + 1 UUAGAUUUUGGCGCGUUCUACGCUCAGCAUCGCGCUGCU--------GGUGACGUAAUGGGCCGUCAAACAGGUUGGCCAAAAUAACAAAAUAACGGCC-------AAAA .....(((((((.((((...(((.(((((......))))--------))))..((.((.(((((.(......).)))))...)).)).....)))))))-------)))) ( -29.80) >DroAna_CAF1 36771 96 + 1 U----UUUUGGCGCGUUCUCAGAUCAGCAUCUCCGAGAUUGAGGCCCGAUGACGUAAUGGGCCGCCAAACAGGUUGCC----------AAAUAACGGCCGGGCCAGAAAG (----(((((((.(..((((((((....))))..))))(((.(((((.((......)))))))..)))...(((((..----------......)))))).)))))))). ( -32.30) >consensus UUAGAUUUUGGCGCGUUCUACGCUCAGCAUCGCGAUGAU_G_G____GGUGACGUAAUGGGCCGUCAAACAGGUUGGCCAAAAUAACAAAAUAACGGCC_______AAAA ......(((((((.((((((((.(((.(...................).)))))))..)))))))))))..(((((..................)))))........... (-17.16 = -17.52 + 0.36)


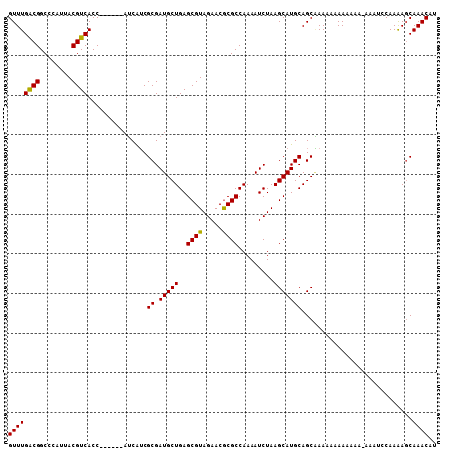
| Location | 16,614,999 – 16,615,100 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.18 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16614999 101 - 22224390 UUUU-------GGCCGUUAUUUUGUUAUUUUGGCCAACCUGUUUGACGGCCCAUUACGUCACCA--UCACCAUCAUCGCGAUGCUGAGCGUAGAACGCGCCAAAAUCUAA ((((-------(((((((.............((((............))))..((((((((.((--((.(.......).)))).))).))))))))).)))))))..... ( -28.10) >DroSim_CAF1 25223 101 - 1 UUUU-------GGCCGUUAUUUUGUUAUUUCGGCCAACCUGUUUGACGGCCCAUUACGUCACCA--UCACCAUCAUCGCGAUGCUGAGCGUAGAACGCGCCAAAAUCUAA ((((-------(((((((.............((((............))))..((((((((.((--((.(.......).)))).))).))))))))).)))))))..... ( -28.40) >DroEre_CAF1 27208 94 - 1 UUUU-------GGCCGCUAUUUUGUAAUUUUGGGCAACCUGUUUGGCGGC-CAUUACGUCACC--------AUCGUCGCGAUGCUGAGCGUAGAACACGCCAAAAUCUAA ...(-------((((((((......(((...((....)).))))))))))-))((((((((.(--------((((...))))).))).)))))................. ( -30.00) >DroYak_CAF1 28889 95 - 1 UUUU-------GGCCGUUAUUUUGUUAUUUUGGCCAACCUGUUUGACGGCCCAUUACGUCACC--------AGCAGCGCGAUGCUGAGCGUAGAACGCGCCAAAAUCUAA ((((-------(((((((.............((((............))))..((((((...(--------((((......))))).)))))))))).)))))))..... ( -29.80) >DroAna_CAF1 36771 96 - 1 CUUUCUGGCCCGGCCGUUAUUU----------GGCAACCUGUUUGGCGGCCCAUUACGUCAUCGGGCCUCAAUCUCGGAGAUGCUGAUCUGAGAACGCGCCAAAA----A ......((((((((((.....)----------)))........(((((........))))).))))))....(((((((........)))))))...........----. ( -33.50) >consensus UUUU_______GGCCGUUAUUUUGUUAUUUUGGCCAACCUGUUUGACGGCCCAUUACGUCACC____C_C_AUCAUCGCGAUGCUGAGCGUAGAACGCGCCAAAAUCUAA ...........(((((((.........................(((((........)))))...............(((........)))...)))).)))......... (-16.66 = -16.18 + -0.48)

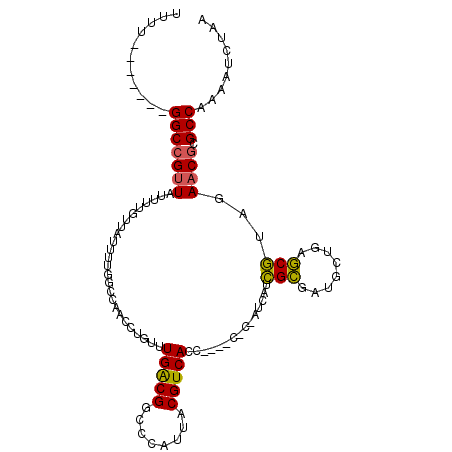
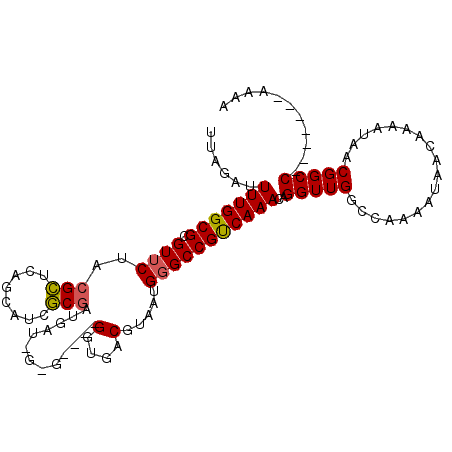
| Location | 16,615,067 – 16,615,158 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 89.65 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16615067 91 + 22224390 AGGUUGGCCAAAAUAACAAAAUAACGGCCAAAAAGCCAUUAGACUUUUAGCCAGAUUGAAAGCCAGCCAGCCAGCCCUUCCCACACAACUU .(((((((.................(((......)))....(.(((((((.....))))))).).)))))))................... ( -20.60) >DroSim_CAF1 25291 91 + 1 AGGUUGGCCGAAAUAACAAAAUAACGGCCAAAAAGCCAUUAGACUUUUAGCCAGAAUGAAAGCCAACCAGCCAGCCCUUCCCACACAACUU .(((((((.(...............(((......)))....(.((((((.......)))))).)...).)))))))............... ( -19.80) >DroEre_CAF1 27269 86 + 1 AGGUUGCCCAAAAUUACAAAAUAGCGGCCAAAAAGCCAUUAGGCUUUUAGCCAGGUCGAA-----GCCAGCCAGCCCUUCCCACACAAAUG .(((((...................(((..(((((((....))))))).))).(((....-----)))...)))))............... ( -21.10) >DroYak_CAF1 28951 86 + 1 AGGUUGGCCAAAAUAACAAAAUAACGGCCAAAAAGCCAUUAGACUUUUAGCCAGCUUGAA-----GCCAGCCAGCCCUUCCCACACAAAUU .(((((((.................(((......)))....(.((((.((....)).)))-----).).)))))))............... ( -19.90) >consensus AGGUUGGCCAAAAUAACAAAAUAACGGCCAAAAAGCCAUUAGACUUUUAGCCAGAUUGAA_____GCCAGCCAGCCCUUCCCACACAAAUU .(((((((.................(((..(((((........))))).))).....(.........).)))))))............... (-13.98 = -14.22 + 0.25)


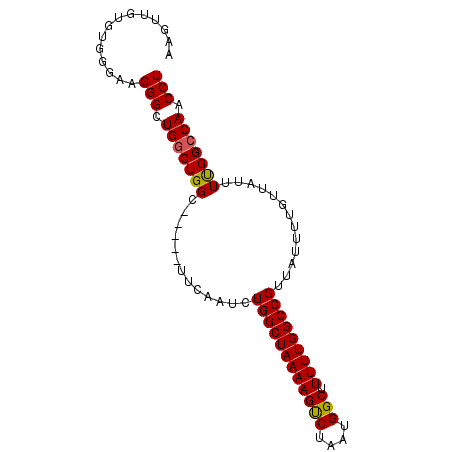
| Location | 16,615,067 – 16,615,158 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 89.65 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -22.84 |
| Energy contribution | -22.72 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16615067 91 - 22224390 AAGUUGUGUGGGAAGGGCUGGCUGGCUGGCUUUCAAUCUGGCUAAAAGUCUAAUGGCUUUUUGGCCGUUAUUUUGUUAUUUUGGCCAACCU .........(((((((.(..(....)..))))))....(((((((((..(((((((((....))))))))....)...))))))))).)). ( -28.70) >DroSim_CAF1 25291 91 - 1 AAGUUGUGUGGGAAGGGCUGGCUGGUUGGCUUUCAUUCUGGCUAAAAGUCUAAUGGCUUUUUGGCCGUUAUUUUGUUAUUUCGGCCAACCU ..((((.((.((((..((.((((..((((((........)))))).))))((((((((....))))))))....))..)))).)))))).. ( -30.80) >DroEre_CAF1 27269 86 - 1 CAUUUGUGUGGGAAGGGCUGGCUGGC-----UUCGACCUGGCUAAAAGCCUAAUGGCUUUUUGGCCGCUAUUUUGUAAUUUUGGGCAACCU .........(((..(((((....)))-----.))..)))(((((((((((....))).))))))))................((....)). ( -25.80) >DroYak_CAF1 28951 86 - 1 AAUUUGUGUGGGAAGGGCUGGCUGGC-----UUCAAGCUGGCUAAAAGUCUAAUGGCUUUUUGGCCGUUAUUUUGUUAUUUUGGCCAACCU .........(((...(((..(((...-----....)))..)))(((((((....)))))))((((((..((......))..)))))).))) ( -27.70) >consensus AAGUUGUGUGGGAAGGGCUGGCUGGC_____UUCAAUCUGGCUAAAAGUCUAAUGGCUUUUUGGCCGUUAUUUUGUUAUUUUGGCCAACCU ..............(((.(((((((.............((((((((((((....))).))))))))).............))))))).))) (-22.84 = -22.72 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:25 2006