
| Sequence ID | X_DroMel_CAF1 |
|---|---|
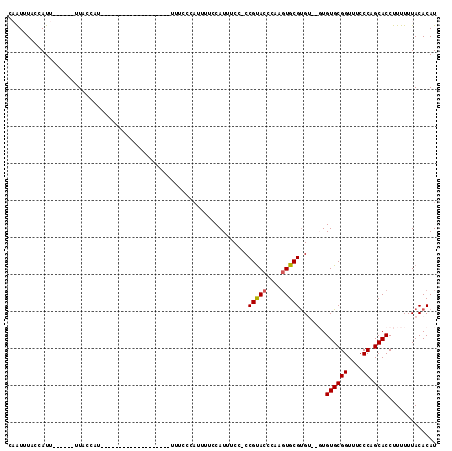
| Location | 16,609,323 – 16,609,414 |
| Length | 91 |
| Max. P | 0.962362 |

| Location | 16,609,323 – 16,609,414 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -12.53 |
| Consensus MFE | -9.88 |
| Energy contribution | -9.92 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16609323 91 + 22224390 CAAUUUACCAUU------UUACCAU-------------------UUUCCCAUUUUCCAUUUCCCCCGUACCCAAGUGCGUGUGUGUGUGCGGUUUCCCAGCACCUUUUUUACACAU ............------.......-------------------.....................(((((....)))))((((((.((((((....)).))))......)))))). ( -14.60) >DroSec_CAF1 21824 86 + 1 CCAUUUUCCAUU------UUACCAU-------------------UUUCCCAUUUUCCAUUUCC-CCGUACCCAAGUGCGUG----UGUGCGGUUUCCCAGCACCUUUUUUACACAU ............------.......-------------------...................-..((((....))))(((----(((((((....)).)))).......)))).. ( -11.51) >DroSim_CAF1 19751 92 + 1 CCAUUUCCCAUUUUCGAUUUACCAU-------------------UUUCCCAUUUUCCAUUUCC-CCGUACCCAAGUGCGUG----UGUGCGGUUUCCCAGCACCUUUUUUACACAU .........................-------------------...................-..((((....))))(((----(((((((....)).)))).......)))).. ( -11.51) >DroEre_CAF1 21832 86 + 1 CAAUUUACCAUU------UUCCCUAUCCAAU---------------------UUUCCAUUUCU-CCGUACCCAACUGCGCGU--GUGUGCGGUUUCCCAGCACCUUUUUUACACAU ............------.............---------------------...........-.((((......)))).((--((((((((....)).)))).......)))).. ( -9.81) >DroYak_CAF1 23302 107 + 1 CAGUUUAGCAUU------UUCCCUUUUCAAUUUCCCAUUUACAAUUUACCAUUUUCCAUUUCC-CCGCACCCAAGUGCGUGU--GUGUGCGGUUUCCCAGCACCUUUUUUACACAU ............------.............................................-.(((((....)))))(((--((((((((....)).)))).......))))). ( -15.21) >consensus CAAUUUACCAUU______UUACCAU___________________UUUCCCAUUUUCCAUUUCC_CCGUACCCAAGUGCGUGU__GUGUGCGGUUUCCCAGCACCUUUUUUACACAU .................................................................(((((....))))).......((((((....)).))))............. ( -9.88 = -9.92 + 0.04)


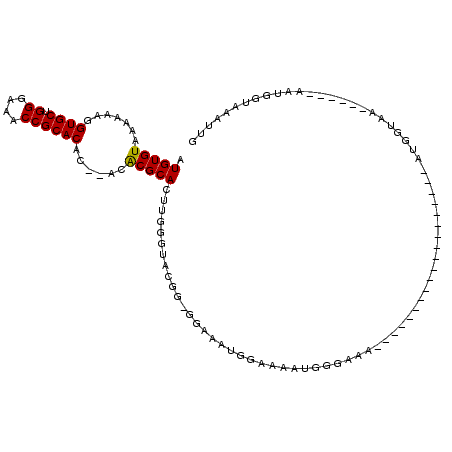
| Location | 16,609,323 – 16,609,414 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16609323 91 - 22224390 AUGUGUAAAAAAGGUGCUGGGAAACCGCACACACACACGCACUUGGGUACGGGGGAAAUGGAAAAUGGGAAA-------------------AUGGUAA------AAUGGUAAAUUG .(((((.......((((.((....))))))......)))))((((....))))...................-------------------.......------............ ( -17.62) >DroSec_CAF1 21824 86 - 1 AUGUGUAAAAAAGGUGCUGGGAAACCGCACA----CACGCACUUGGGUACGG-GGAAAUGGAAAAUGGGAAA-------------------AUGGUAA------AAUGGAAAAUGG .(((((.......((((.((....)))))).----.)))))((((....)))-)..................-------------------.......------............ ( -16.00) >DroSim_CAF1 19751 92 - 1 AUGUGUAAAAAAGGUGCUGGGAAACCGCACA----CACGCACUUGGGUACGG-GGAAAUGGAAAAUGGGAAA-------------------AUGGUAAAUCGAAAAUGGGAAAUGG .(((((.......((((.((....)))))).----.)))))((((....)))-)..................-------------------......................... ( -16.00) >DroEre_CAF1 21832 86 - 1 AUGUGUAAAAAAGGUGCUGGGAAACCGCACAC--ACGCGCAGUUGGGUACGG-AGAAAUGGAAA---------------------AUUGGAUAGGGAA------AAUGGUAAAUUG .(((((.......((((.((....))))))..--..)))))...........-...........---------------------.............------............ ( -14.80) >DroYak_CAF1 23302 107 - 1 AUGUGUAAAAAAGGUGCUGGGAAACCGCACAC--ACACGCACUUGGGUGCGG-GGAAAUGGAAAAUGGUAAAUUGUAAAUGGGAAAUUGAAAAGGGAA------AAUGCUAAACUG .(((((.......((((.((....))))))))--)))(((((....))))).-.............................................------............ ( -21.31) >consensus AUGUGUAAAAAAGGUGCUGGGAAACCGCACAC__ACACGCACUUGGGUACGG_GGAAAUGGAAAAUGGGAAA___________________AUGGUAA______AAUGGUAAAUUG .(((((.......((((.((....))))))......)))))........................................................................... (-15.30 = -15.14 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:17 2006