
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,592,419 – 16,592,514 |
| Length | 95 |
| Max. P | 0.872469 |

| Location | 16,592,419 – 16,592,514 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
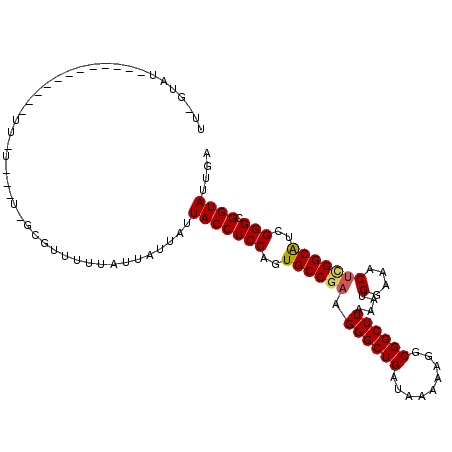
>X_DroMel_CAF1 16592419 95 + 22224390 UU-UUAU------------UUUU---UGGCAUUUUUAUUAUUAUUACCUCCAGUGCCGAAGGGCUGAUAAAGAGGCGGCUUAAAUGGAAACUUGGCAUCGGGCGGUAUCGA ..-....------------....---..................(((((((.(((((((.((((((.........))))))....(....)))))))).))).)))).... ( -23.70) >DroPse_CAF1 6552 90 + 1 UUUGUAU------------UU---------UUUUGUAUUAUUAUUACCUCCAGUGCCGAAGGGCUGAUAAUAAGGCGGCUUAAAUGGAAACUCGGCAUCGGGCGGUAUUGA .......------------..---------..........(((.(((((((.(((((((.((((((.........))))))....(....)))))))).))).)))).))) ( -26.40) >DroEre_CAF1 5204 93 + 1 UU-UUAU-------------U-U---UUGCGUUUUUAUUAUUAUUACCUCCAGUGCCGAAGGGCUGAUAAAGAGGCGGCUUAAAUGGAAACUCGGCAUCGGGCGGUAUCGA ..-....-------------.-.---..................(((((((.(((((((.((((((.........))))))....(....)))))))).))).)))).... ( -25.80) >DroWil_CAF1 5811 80 + 1 -------------------------------GUUUUGUUUUUAUUACCUCCAGUGCCGAAGGGCUGAUAAAAAGGCGGCUUAUAUGGAAACGAGGCAUCGGGCGGUAUUGA -------------------------------.........(((.(((((((.(((((...((((((.........))))))...((....)).))))).))).)))).))) ( -23.80) >DroMoj_CAF1 5382 104 + 1 UU-GCAGUGUGAGAAGCGAA--U---UUGCG-UAUUUUUAUUAUUACCUCCAGUGCCGAAGGGCUGAUAAUAAGGCGGCUUAAAUGGAAACUCGGCAUCGGGCGGUAUUGA ..-.....((((((((((..--.---...))-).)))))))...(((((((.(((((((.((((((.........))))))....(....)))))))).))).)))).... ( -29.70) >DroAna_CAF1 5198 104 + 1 UU-GUAUAUU-----GUGUUU-UUGAUGGCUUUUCAAAGAUUAUUACCUCCAGUGCCGAAGGGCUGAUAAUAAGGCGGCUUAAAUGGAAACUCGGCGUCGGGCGGUAUUGA ..-.......-----..((((-((((.......))))))))...(((((((..((((((.((((((.........))))))....(....)))))))..))).)))).... ( -29.40) >consensus UU_GUAU____________UU_U___U_GCGUUUUUAUUAUUAUUACCUCCAGUGCCGAAGGGCUGAUAAAAAGGCGGCUUAAAUGGAAACUCGGCAUCGGGCGGUAUUGA ............................................(((((((..((((((.((((((.........))))))....(....)))))))..))).)))).... (-24.08 = -24.17 + 0.08)

| Location | 16,592,419 – 16,592,514 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -17.05 |
| Consensus MFE | -15.13 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16592419 95 - 22224390 UCGAUACCGCCCGAUGCCAAGUUUCCAUUUAAGCCGCCUCUUUAUCAGCCCUUCGGCACUGGAGGUAAUAAUAAUAAAAAUGCCA---AAAA------------AUAA-AA ....((((..(((.((((..((((......)))).((..........)).....)))).))).))))..................---....------------....-.. ( -14.60) >DroPse_CAF1 6552 90 - 1 UCAAUACCGCCCGAUGCCGAGUUUCCAUUUAAGCCGCCUUAUUAUCAGCCCUUCGGCACUGGAGGUAAUAAUAAUACAAAA---------AA------------AUACAAA ....((((..(((.(((((((........((((....))))..........))))))).))).))))..............---------..------------....... ( -18.57) >DroEre_CAF1 5204 93 - 1 UCGAUACCGCCCGAUGCCGAGUUUCCAUUUAAGCCGCCUCUUUAUCAGCCCUUCGGCACUGGAGGUAAUAAUAAUAAAAACGCAA---A-A-------------AUAA-AA ....((((..(((.(((((((.........(((......))).........))))))).))).))))..................---.-.-------------....-.. ( -18.27) >DroWil_CAF1 5811 80 - 1 UCAAUACCGCCCGAUGCCUCGUUUCCAUAUAAGCCGCCUUUUUAUCAGCCCUUCGGCACUGGAGGUAAUAAAAACAAAAC------------------------------- ....((((..(((.((((..((((......)))).((..........)).....)))).))).)))).............------------------------------- ( -14.20) >DroMoj_CAF1 5382 104 - 1 UCAAUACCGCCCGAUGCCGAGUUUCCAUUUAAGCCGCCUUAUUAUCAGCCCUUCGGCACUGGAGGUAAUAAUAAAAAUA-CGCAA---A--UUCGCUUCUCACACUGC-AA ....((((..(((.(((((((........((((....))))..........))))))).))).))))............-.((..---.--...))............-.. ( -19.57) >DroAna_CAF1 5198 104 - 1 UCAAUACCGCCCGACGCCGAGUUUCCAUUUAAGCCGCCUUAUUAUCAGCCCUUCGGCACUGGAGGUAAUAAUCUUUGAAAAGCCAUCAA-AAACAC-----AAUAUAC-AA ....((((..(((..((((((........((((....))))..........))))))..))).))))......(((((.......))))-).....-----.......-.. ( -17.07) >consensus UCAAUACCGCCCGAUGCCGAGUUUCCAUUUAAGCCGCCUUAUUAUCAGCCCUUCGGCACUGGAGGUAAUAAUAAUAAAAAAGC_A___A_AA____________AUAC_AA ....((((..(((.((((((((((......)))).((..........))...)))))).))).))))............................................ (-15.13 = -15.80 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:12 2006