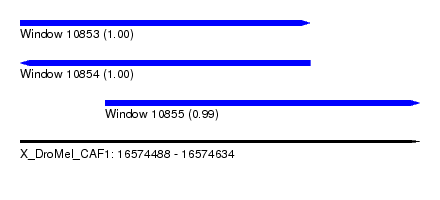
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,574,488 – 16,574,634 |
| Length | 146 |
| Max. P | 0.999644 |

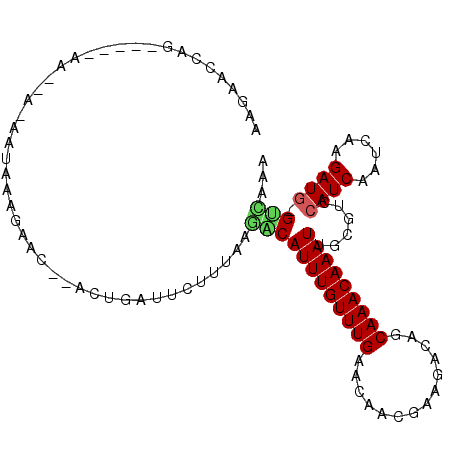
| Location | 16,574,488 – 16,574,594 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.43 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16574488 106 + 22224390 AAGAACCAGUAAAAAAAAAAAAUAAAGAACUCACUGAUUCUUUCAGACAUUUGUUUGAACAACGAAGAUAGCAAACAAAUUGCGUCCAUCAAUCAAGAUG-GUCAAA (((((.((((......................)))).)))))((((((....))))))............((((.....))))(.(((((......))))-).)... ( -20.15) >DroSim_CAF1 14698 101 + 1 AAGAACCAG-----AAAAAUAAUAAAGAACCAACUGAUUCUUUCAGACAUUUGUUUGAACAACGAAGACAGCAAACAAAUUGCGCCCAUCAAUCAAGAUG-GUCAAA (((((.(((-----...................))).)))))((((((....))))))........(((.((((.....))))...((((......))))-)))... ( -18.11) >DroEre_CAF1 14133 82 + 1 AAGAACCA------------------------ACUGAUUCUUUAAAGCAUUUGUUUGAACAACAAAGACAGCAAACAAAUUGCGUCCAUCAAUCAAGAUG-GUUAAA ...(((((------------------------.((((((.......(((((((((((..............)))))))).))).......)))).)).))-)))... ( -17.58) >DroYak_CAF1 15227 97 + 1 AAGAACCAG-----AA----AAUAAAGAACCCACUGAUUCUUGAAGGCAUUUGUUUGAACAACAAAGACAGCAAACAAAUCGCGUCCAUCAAUCAAGAUG-GCUAAA (((((.(((-----..----.............))).)))))....(((((((((((..............))))))))).))(.(((((......))))-)).... ( -18.80) >DroAna_CAF1 23972 94 + 1 AAGAACCAU-----AAGGAGAUUGAAUA-------AAUUCUUCUGGCCAUUUGUUUGCUCAG-GCAGACAGCAAACAAAUGGCUCCCAUCAAUCAAGAUAUGUAAAA ......(((-----(((((((.(.....-------.).))))))(((((((((((((((...-......)))))))))))))))..............))))..... ( -27.10) >consensus AAGAACCAG_____AA__A_AAUAAAGAAC__ACUGAUUCUUUAAGACAUUUGUUUGAACAACGAAGACAGCAAACAAAUUGCGUCCAUCAAUCAAGAUG_GUCAAA .............................................((((((((((((..............)))))))))......((((......)))).)))... ( -8.68 = -8.72 + 0.04)


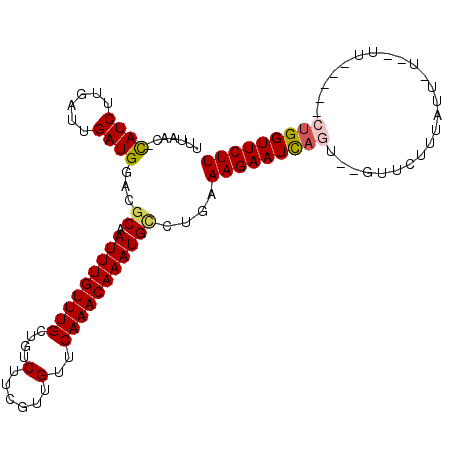
| Location | 16,574,488 – 16,574,594 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16574488 106 - 22224390 UUUGAC-CAUCUUGAUUGAUGGACGCAAUUUGUUUGCUAUCUUCGUUGUUCAAACAAAUGUCUGAAAGAAUCAGUGAGUUCUUUAUUUUUUUUUUUUACUGGUUCUU .....(-((((......)))))..(((.((((((((....(......)..)))))))))))....(((((((((((((................))))))))))))) ( -25.59) >DroSim_CAF1 14698 101 - 1 UUUGAC-CAUCUUGAUUGAUGGGCGCAAUUUGUUUGCUGUCUUCGUUGUUCAAACAAAUGUCUGAAAGAAUCAGUUGGUUCUUUAUUAUUUUU-----CUGGUUCUU ..((.(-((((......)))))...))(((((((((....(......)..)))))))))......(((((((((..(((........)))...-----))))))))) ( -22.20) >DroEre_CAF1 14133 82 - 1 UUUAAC-CAUCUUGAUUGAUGGACGCAAUUUGUUUGCUGUCUUUGUUGUUCAAACAAAUGCUUUAAAGAAUCAGU------------------------UGGUUCUU .....(-((((......)))))..(((.((((((((....(......)..)))))))))))....(((((((...------------------------.))))))) ( -20.90) >DroYak_CAF1 15227 97 - 1 UUUAGC-CAUCUUGAUUGAUGGACGCGAUUUGUUUGCUGUCUUUGUUGUUCAAACAAAUGCCUUCAAGAAUCAGUGGGUUCUUUAUU----UU-----CUGGUUCUU ....((-((((......))))).)((.(((((((((....(......)..)))))))))))....(((((((((..(((.....)))----..-----))))))))) ( -25.40) >DroAna_CAF1 23972 94 - 1 UUUUACAUAUCUUGAUUGAUGGGAGCCAUUUGUUUGCUGUCUGC-CUGAGCAAACAAAUGGCCAGAAGAAUU-------UAUUCAAUCUCCUU-----AUGGUUCUU .....((((....((((((...(.((((((((((((((......-...))))))))))))))).........-------...))))))....)-----)))...... ( -27.96) >consensus UUUAAC_CAUCUUGAUUGAUGGACGCAAUUUGUUUGCUGUCUUCGUUGUUCAAACAAAUGCCUGAAAGAAUCAGU__GUUCUUUAUU_U__UU_____CUGGUUCUU .......((((......))))...((.(((((((((....(......)..)))))))))))....(((((((((........................))))))))) (-14.10 = -14.42 + 0.32)


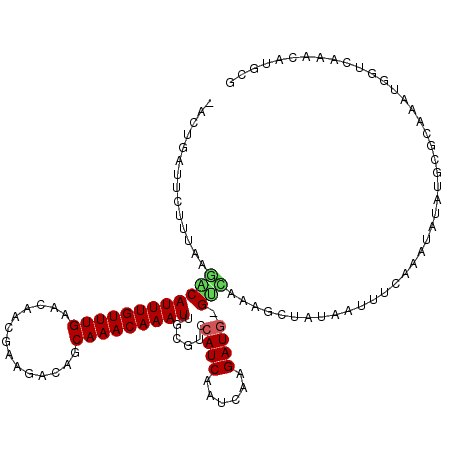
| Location | 16,574,519 – 16,574,634 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.88 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.34 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16574519 115 + 22224390 CACUGAUUCUUUCAGACAUUUGUUUGAACAACGAAGAUAGCAAACAAAUUGCGUCCAUCAAUCAAGAUG-GUCAAAGCUAUAGUUUCAAGUAUAUGCACACAUGUUCAAACAUUCU ..((((.....)))).....((((((((((.........((((.....))))(.(((((......))))-).)...((((((.(....).)))).)).....)))))))))).... ( -28.50) >DroSim_CAF1 14724 115 + 1 AACUGAUUCUUUCAGACAUUUGUUUGAACAACGAAGACAGCAAACAAAUUGCGCCCAUCAAUCAAGAUG-GUCAAAGCUAUAAUUUCAAGUAUAUGCACAACCGGUCAAACAUUCC ..((((.....)))).....(((((((....((..(((.((((.....))))...((((......))))-)))...((((((........)))).)).....)).))))))).... ( -23.90) >DroEre_CAF1 14141 114 + 1 -ACUGAUUCUUUAAAGCAUUUGUUUGAACAACAAAGACAGCAAACAAAUUGCGUCCAUCAAUCAAGAUG-GUUAAAGCUAUAAUUUCAAAUAAAUGCGCACAUGGUCAAAUAUGCG -..............(((((((((((((...........((((.....))))..(((((......))))-).............)))))))))))))(((.((......)).))). ( -27.10) >DroYak_CAF1 15249 115 + 1 CACUGAUUCUUGAAGGCAUUUGUUUGAACAACAAAGACAGCAAACAAAUCGCGUCCAUCAAUCAAGAUG-GCUAAAGCUAAAAUUUCAAAUAUAUGCGCAAAUGGUCAAGUGUGCG ((((((((.(((...((((.((((((((...........((.........))(.(((((......))))-))............)))))))).)))).)))..)))).)))).... ( -24.10) >DroAna_CAF1 23995 108 + 1 ----AAUUCUUCUGGCCAUUUGUUUGCUCAG-GCAGACAGCAAACAAAUGGCUCCCAUCAAUCAAGAUAUGUAAAAACUAUACUUUAAUCCAUGUUUGCUUAUACUAAAAAAA--- ----.........(((((((((((((((...-......)))))))))))))))...........(((((((((((........))))...)))))))................--- ( -23.30) >consensus _ACUGAUUCUUUAAGACAUUUGUUUGAACAACGAAGACAGCAAACAAAUUGCGUCCAUCAAUCAAGAUG_GUCAAAGCUAUAAUUUCAAAUAUAUGCGCAAAUGGUCAAACAUGCG ..............((((((((((((..............)))))))))......((((......)))).)))........................................... ( -8.68 = -8.72 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:05 2006