
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,566,385 – 16,566,487 |
| Length | 102 |
| Max. P | 0.756133 |

| Location | 16,566,385 – 16,566,487 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.54 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -29.41 |
| Energy contribution | -28.42 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16566385 102 + 22224390 GCCUCGGAAGAUUCG-GACUGGAUGGGAAAGCCCCUUGGG-UUUGGGGCCCAUUACAGUGGAAUCUCGCCGGGGAAACCUUUGCGAUUUUUC----CUAUUUAGU--UUU ...((....))...(-(((((((((((((((((((.....-...)))))((((....))))....((((.(((....)))..))))..))))----)))))))))--)). ( -42.70) >DroSec_CAF1 6192 102 + 1 GCCUCGGAAGAUUCU-GAUUGGAUGGGAAAGCCCCUUGGG-UUUGGGGCCCAUUGCAGUGGAAUCUUGCCGGGGAAACCUUUGCGAUUUUUC----CUAUUUAGU--UUU ...((....))....-(((((((((((((((((((.....-...)))))((((....))))....((((.(((....)))..))))..))))----)))))))))--).. ( -37.70) >DroSim_CAF1 6590 102 + 1 GCCCCGGAAGAUUCG-GACUGGAUGGGAAAGCCCCUUGGG-UUUGGGGCCCAUUACAGUGGAAUCUUGCCGGGGAAACCUUUGCGAUUUUUC----CUAUUUAGU--UUU .(((((((((((((.-(.(((((((((.((((((...)))-)))....)))))).)))).))))))).))))))..................----.........--... ( -41.10) >DroEre_CAF1 6278 100 + 1 GCCUCGGAAGAUUCG-GACUGAAUG-GAAAGCCCCUUGGG-UUUGGGGCCCAUUACAGUGGAAUCUCGUCGGGGAAACCUUUGCGAUU-UUC----CUAUUUGGU--UUU (((..(((((((((.-(.(((((((-(...(((((.....-...)))))))))).)))).)))))((((.(((....)))..))))..-)))----).....)))--... ( -38.50) >DroYak_CAF1 7114 102 + 1 GCCCUGAAAGAUUCG-GACUGGAUGGGAAAACCCCUUGGG-UUUGGGGCCCAUUACAGUGGAAUCUUGUUGGGGAAACCUUUGCGAUUUUUC----CUAUUUAGU--UUU ...((((.....)))-)((((((((((((((((((.....-...)))).((((....))))....((((.(((....)))..)))).)))))----)))))))))--... ( -35.20) >DroAna_CAF1 11993 110 + 1 GCCUCGGCGGACUCGGGACUGGAUGGCCAAUCCUCUAGGGAUUUGGGAUCUAUCGCAGUGGAGUGCCGCCGGGAAGACCUGCGCGAUUUUUCUAUUCUAUUUAGAUUCUU ..((((((((((((...((((((((((((((((.....))).))))...))))).)))).)))).))))))))...........((....((((.......)))).)).. ( -38.10) >consensus GCCUCGGAAGAUUCG_GACUGGAUGGGAAAGCCCCUUGGG_UUUGGGGCCCAUUACAGUGGAAUCUCGCCGGGGAAACCUUUGCGAUUUUUC____CUAUUUAGU__UUU .((((((..(((((...((((((((((....((((.........)))))))))).)))).)))))...)))))).................................... (-29.41 = -28.42 + -0.99)

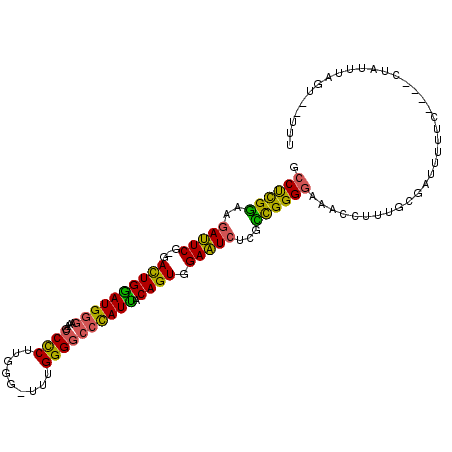
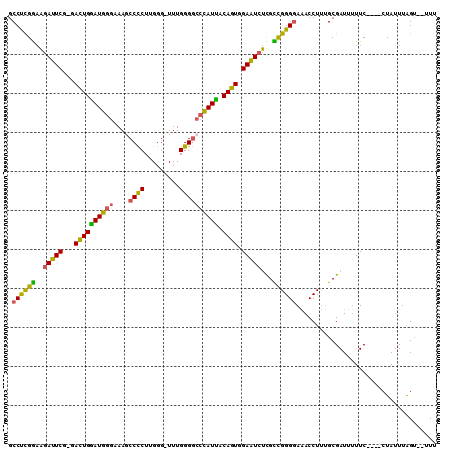
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:02 2006