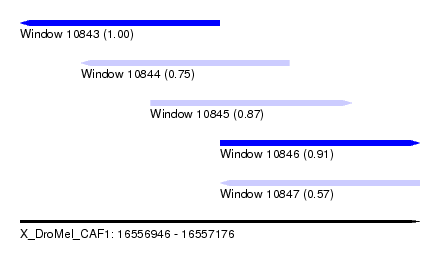
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,556,946 – 16,557,176 |
| Length | 230 |
| Max. P | 0.999759 |

| Location | 16,556,946 – 16,557,061 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -22.52 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16556946 115 - 22224390 UGCUGCCACCUGGGUAGCCAAGCGAAACCGUAGCCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGUAAAUUCUA-AAACUUCUCUACCA---- .((((((.....)))))).....(((((.(((.......))).)))))....(((.(.(((.........((((....))))((....))))).).)))..-..............---- ( -21.30) >DroSec_CAF1 12516 119 - 1 UGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGUAAUUUCGA-AAACUUCCCUACAAUAUU .((((((.....)))))).....(((((.(((.......))).)))))...((((((.(((.........((((....))))((....))))).)))))).-.................. ( -26.50) >DroSim_CAF1 9864 119 - 1 UGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAAACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGUAAUUUCGA-AAACUUCUCUACAAUAUU .((((((.....)))))).....(((((.((.........)).)))))...((((((.(((.........((((....))))((....))))).)))))).-.................. ( -24.90) >DroEre_CAF1 11039 119 - 1 UGCUGCCACCUGGGUAGCCAACCGAAACCGUAACCAUUAUACCGUUUCAACGGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAAGGUUAUUUCAA-AAACAUCUCUACUAUAAU .((((((.....)))))).....(((((.(((.......))).)))))....(((((((((...(((...((((....)))).)))....)))))))))..-.................. ( -27.90) >DroYak_CAF1 12435 113 - 1 UGCUGCCACCUGGGUAGCCAACCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAAGGUAAUUUCGAGAAACAUUUCUA------- .((((((.....)))))).....(((((.(((.......))).)))))...((((((.(((...(((...((((....)))).)))....))).))))))((((....)))).------- ( -26.80) >consensus UGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGUAAUUUCGA_AAACUUCUCUACAAUA_U .((((((.....)))))).....(((((.(((.......))).)))))....(((((.(((...(((...((((....)))).)))....))).)))))..................... (-22.52 = -22.92 + 0.40)



| Location | 16,556,981 – 16,557,101 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -23.81 |
| Consensus MFE | -22.87 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16556981 120 - 22224390 GCAUCUGGAGGAACAUUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAAGCGAAACCGUAGCCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGU .........((........((((((.....((((.......((((((.....)))))).....(((((.(((.......))).)))))....))))......))))))........)).. ( -24.09) >DroSec_CAF1 12555 120 - 1 GCAUCUGGAGGAACAUUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGU .........((........((((((.....((((.......((((((.....)))))).....(((((.(((.......))).)))))....))))......))))))........)).. ( -23.29) >DroSim_CAF1 9903 120 - 1 GCAUCUGGAGGAACAUUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAAACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGU .........((........((((((.....((((.......((((((.....)))))).....(((((.((.........)).)))))....))))......))))))........)).. ( -21.69) >DroEre_CAF1 11078 120 - 1 GCAUCUGGAGGAACACUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAACCGAAACCGUAACCAUUAUACCGUUUCAACGGAAAUAACCGAAUCAAUUUUACAACCGU .........((........((((((.((((((((.......((((((.....)))))).....(((((.(((.......))).)))))...))))))))...))))))........)).. ( -26.69) >DroYak_CAF1 12468 120 - 1 GCAUCUGGAGGAACACUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAACCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGU .........((........((((((.....((((.......((((((.....)))))).....(((((.(((.......))).)))))....))))......))))))........)).. ( -23.29) >consensus GCAUCUGGAGGAACAUUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGU .........((........((((((.....((((.......((((((.....)))))).....(((((.(((.......))).)))))....))))......))))))........)).. (-22.87 = -23.07 + 0.20)



| Location | 16,557,021 – 16,557,137 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -27.30 |
| Energy contribution | -27.74 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16557021 116 + 22224390 AUAAUGGCUACGGUUUCGCUUGGCUACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUGAGUAUAG .......(((((((.((((((((....))))))((.((((.(((((((.........))))))).))))))....)).)))))))(((((.((...----...)).)))))......... ( -33.60) >DroSec_CAF1 12595 116 + 1 AUAAUGGUUACGGUUUCGCUUGGCUACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUGAGUAUAG ........((((((.((((((((....))))))((.((((.(((((((.........))))))).))))))....)).))))))((((((.((...----...)).))))))........ ( -31.90) >DroSim_CAF1 9943 116 + 1 UUAAUGGUUACGGUUUCGCUUGGCUACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAAGUGCCCCAAAC----AGCUCAGGUACUGAGUAUAG .....(((((((((.((((((((....))))))((.((((.(((((((.........))))))).))))))....)).))))))...)))......----.((((((...)))))).... ( -31.30) >DroEre_CAF1 11118 120 + 1 AUAAUGGUUACGGUUUCGGUUGGCUACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAGUGUUCCUCCAGAUGCUGAAGGUGCCUCAUACAUAUAGCUGAAGUACUGAGAAUAG ..........(((((((((((((((((....)))))((((.(((((((.........))))))).))))(((.(((...))).))).............))))))))..))))....... ( -27.20) >DroYak_CAF1 12508 116 + 1 AUAAUGGUUACGGUUUCGGUUGGCUACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAGUGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUAAGAAUAG ........((((((.(((((.....)))..((.((.((((.(((((((.........))))))).)))))).)).)).))))))((((((.((...----...)).))))))........ ( -31.80) >consensus AUAAUGGUUACGGUUUCGCUUGGCUACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC____AGCUGAGGUACUGAGUAUAG ........((((((.((...(((....)))((.((.((((.(((((((.........))))))).)))))).)).)).))))))((((((.((..........)).))))))........ (-27.30 = -27.74 + 0.44)



| Location | 16,557,061 – 16,557,176 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.46 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16557061 115 + 22224390 AUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAA-AAACUUUGUAGCUGUCGCAAAU .(((((((.........)))))))(((((..((.((((((((((((((((.((...----...)).))))))...)))))))))).)))))))....-..........((....)).... ( -26.80) >DroSec_CAF1 12635 115 + 1 AUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAA-AAACUUUGUAGCUGUCGCAAAU .(((((((.........)))))))(((((..((.((((((((((((((((.((...----...)).))))))...)))))))))).)))))))....-..........((....)).... ( -26.80) >DroSim_CAF1 9983 116 + 1 AUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAAGUGCCCCAAAC----AGCUCAGGUACUGAGUAUAGCAUCUAGAAACAUUUAAAAAACUUUGUAGCUGUCGCAAAU .(((((((.........)))))))(((((..((.((((((((((((((((......----......))))))...)))))))))).)))))))...............((....)).... ( -26.10) >DroEre_CAF1 11158 119 + 1 AUAUUUGAAAAUAAUAAUCAAAUAGUGUUCCUCCAGAUGCUGAAGGUGCCUCAUACAUAUAGCUGAAGUACUGAGAAUAGCGUCUACAAUUAUUUUA-AAACAUUGUACUUAUCGCCAAU .(((((((.........)))))))(((.......((((((((..(((((.(((..........))).))))).....))))))))(((((.......-....)))))......))).... ( -19.70) >DroYak_CAF1 12548 115 + 1 AUAUUUGAAAAUAAUAAUCAAAUAGUGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUAAGAAUAGCAUUUACAAUUAUUUAA-AAGCAUUGUUCUUAUCGUAAAG .(((((((.........)))))))..((.((((.....(((((.((....))..))----))).)))).))((((((((((.((((........)))-).))..))))))))........ ( -22.20) >consensus AUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC____AGCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAA_AAACUUUGUAGCUGUCGCAAAU .(((((((.........)))))))..........((((((((((((((((.((..........)).))))))...))))))))))................................... (-19.94 = -20.46 + 0.52)

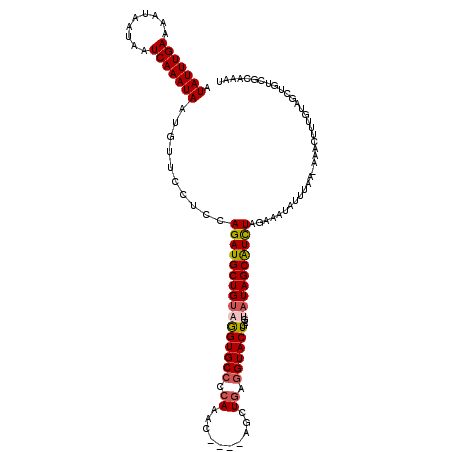

| Location | 16,557,061 – 16,557,176 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -18.54 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16557061 115 - 22224390 AUUUGCGACAGCUACAAAGUUU-UUAAAUAUUUCUAGAUGCUAUACUCAGUACCUCAGCU----GUUUGGGGCACCUACAGCAUCUGGAGGAACAUUAUUUGAUUAUUAUUUUCAAAUAU ....((....))..........-.......((((((((((((.((....((.((((((..----..)))))).)).)).)))))))))))).....(((((((.........))))))). ( -26.20) >DroSec_CAF1 12635 115 - 1 AUUUGCGACAGCUACAAAGUUU-UUAAAUAUUUCUAGAUGCUAUACUCAGUACCUCAGCU----GUUUGGGGCACCUACAGCAUCUGGAGGAACAUUAUUUGAUUAUUAUUUUCAAAUAU ....((....))..........-.......((((((((((((.((....((.((((((..----..)))))).)).)).)))))))))))).....(((((((.........))))))). ( -26.20) >DroSim_CAF1 9983 116 - 1 AUUUGCGACAGCUACAAAGUUUUUUAAAUGUUUCUAGAUGCUAUACUCAGUACCUGAGCU----GUUUGGGGCACUUACAGCAUCUGGAGGAACAUUAUUUGAUUAUUAUUUUCAAAUAU ....((....))..............((((((((((((((((...(((((...)))))..----((..(.....)..))))))))))))..))))))((((((.........)))))).. ( -26.50) >DroEre_CAF1 11158 119 - 1 AUUGGCGAUAAGUACAAUGUUU-UAAAAUAAUUGUAGACGCUAUUCUCAGUACUUCAGCUAUAUGUAUGAGGCACCUUCAGCAUCUGGAGGAACACUAUUUGAUUAUUAUUUUCAAAUAU ..(((((.....((((((....-.......))))))..))))).(((((.(((...........))))))))..(((((((...))))))).....(((((((.........))))))). ( -23.60) >DroYak_CAF1 12548 115 - 1 CUUUACGAUAAGAACAAUGCUU-UUAAAUAAUUGUAAAUGCUAUUCUUAGUACCUCAGCU----GUUUGGGGCACCUACAGCAUCUGGAGGAACACUAUUUGAUUAUUAUUUUCAAAUAU ........((((((....((.(-(((........)))).))..))))))((.((((.(((----((..((....)).))))).....)))).))..(((((((.........))))))). ( -24.70) >consensus AUUUGCGACAGCUACAAAGUUU_UUAAAUAUUUCUAGAUGCUAUACUCAGUACCUCAGCU____GUUUGGGGCACCUACAGCAUCUGGAGGAACAUUAUUUGAUUAUUAUUUUCAAAUAU ...............................(((((((((((.......((.((((((........)))))).))....)))))))))))......(((((((.........))))))). (-18.54 = -19.42 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:58 2006