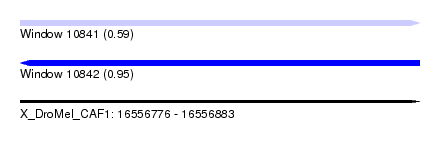
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,556,776 – 16,556,883 |
| Length | 107 |
| Max. P | 0.950978 |

| Location | 16,556,776 – 16,556,883 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16556776 107 + 22224390 ACCGAGCAGCACGACAAGGGCAUCGUGGAUAUGGUG---GCCACCACCGGAGACUCCAACCGGGACGCCAUGCUCAUGCUGCUCCAGAUGAUGUGAGUAUACA------CAUUUGA ...((((((((.((((..(((.((.(((....((((---(...)))))((.....))..))).)).))).)).)).))))))))((((((.((((....))))------)))))). ( -44.00) >DroSec_CAF1 12308 110 + 1 ACCGAGCAGCACGACAAGGGCAUCGUGGAUAUGAUG---GGCACCACCGGAGACUC---CCGGGACGCCAUGCUCAUGCUGCUCCAGAUUAUGUGAGUAUGCAUAACAACAUUUGA ...((((((((.((((....((((((....))))))---(((..(.((((......---)))))..))).)).)).))))))))((((((((((......))))).....))))). ( -36.70) >DroSim_CAF1 9656 110 + 1 ACCGAGCAGCACGACAAGGGCAUCGUGGAUAUGAUG---GGCACCACCGGAGACUC---CCGGGACGCCAUGCUCAUGCUGCUCCAGAUGAUGUGAGUAUACAUGACACCAUUUUA ...((((((((.((((....((((((....))))))---(((..(.((((......---)))))..))).)).)).)))))))).(((((.(((.(.......).))).))))).. ( -36.70) >DroEre_CAF1 10873 99 + 1 AGCGAACAGCACGACAAGGGCAUUGUGGAUAUGGUG---GGCGCCACCGGGGACUCCACCCGGGACACCAUGCUCAUGCUGCUCCAGAUGAUGUGAGUACAC-------------- .((.....)).......((((((.(((....((((.---...))))(((((.......)))))..))).)))))).((.(((((((.....)).))))))).-------------- ( -34.70) >DroYak_CAF1 12245 99 + 1 AGCGAGCAGCACGACAAGGGCAUCGUGGAUAUGGUG---GGCGCCACUGGAGACGCCUCCCGGGACACCAUGCUCAUGCUGCUCCAGAUGAUGUGAGUAUAU-------------- .(.((((((((......((((((.(((....(((..---((((.(......).))))..)))...))).)))))).))))))))).................-------------- ( -36.90) >DroAna_CAF1 16907 110 + 1 GCAGAUCAGCACGACAAGGGCAUCGUGGACAUUGUGCGGGGAGUGACGGGGGAUGCGCCACGGGACACCAUACUCAUGAUGCUCCAAAUGCUGUAAGUAUGCAUA------UUUCA (((...(((((......(((((((((((.((((.(.((........)).).))))..))))((....))........)))))))....)))))......)))...------..... ( -34.80) >consensus ACCGAGCAGCACGACAAGGGCAUCGUGGAUAUGGUG___GGCACCACCGGAGACUCC_CCCGGGACACCAUGCUCAUGCUGCUCCAGAUGAUGUGAGUAUACA________UUU_A ...((((((((......((((((.(((....(((((.....)))))((((.........))))..))).)))))).))))))))................................ (-23.94 = -24.47 + 0.53)


| Location | 16,556,776 – 16,556,883 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -28.11 |
| Energy contribution | -28.94 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16556776 107 - 22224390 UCAAAUG------UGUAUACUCACAUCAUCUGGAGCAGCAUGAGCAUGGCGUCCCGGUUGGAGUCUCCGGUGGUGGC---CACCAUAUCCACGAUGCCCUUGUCGUGCUGCUCGGU ....(((------((......)))))......(((((((((((.((.((((((..((..((.....))(((((...)---))))....))..))))))..)))))))))))))... ( -44.60) >DroSec_CAF1 12308 110 - 1 UCAAAUGUUGUUAUGCAUACUCACAUAAUCUGGAGCAGCAUGAGCAUGGCGUCCCGG---GAGUCUCCGGUGGUGCC---CAUCAUAUCCACGAUGCCCUUGUCGUGCUGCUCGGU .........((((((........))))))...(((((((((((.((.((((((.(((---(....))))((((....---........))))))))))..)))))))))))))... ( -41.20) >DroSim_CAF1 9656 110 - 1 UAAAAUGGUGUCAUGUAUACUCACAUCAUCUGGAGCAGCAUGAGCAUGGCGUCCCGG---GAGUCUCCGGUGGUGCC---CAUCAUAUCCACGAUGCCCUUGUCGUGCUGCUCGGU ....(((((((...(....)..)))))))...(((((((((((.((.((((((.(((---(....))))((((....---........))))))))))..)))))))))))))... ( -43.40) >DroEre_CAF1 10873 99 - 1 --------------GUGUACUCACAUCAUCUGGAGCAGCAUGAGCAUGGUGUCCCGGGUGGAGUCCCCGGUGGCGCC---CACCAUAUCCACAAUGCCCUUGUCGUGCUGUUCGCU --------------(((....)))........((((((((((((((((((((((((((.......))))).))))))---.............)))).....)))))))))))... ( -38.51) >DroYak_CAF1 12245 99 - 1 --------------AUAUACUCACAUCAUCUGGAGCAGCAUGAGCAUGGUGUCCCGGGAGGCGUCUCCAGUGGCGCC---CACCAUAUCCACGAUGCCCUUGUCGUGCUGCUCGCU --------------..................(((((((((((((((.(((....((..((((((......))))))---..)).....))).)))).....)))))))))))... ( -42.70) >DroAna_CAF1 16907 110 - 1 UGAAA------UAUGCAUACUUACAGCAUUUGGAGCAUCAUGAGUAUGGUGUCCCGUGGCGCAUCCCCCGUCACUCCCCGCACAAUGUCCACGAUGCCCUUGUCGUGCUGAUCUGC .....------............((((((..((.(((((.((..(((.((((...((((((.......)))))).....)))).)))..)).))))))).....))))))...... ( -31.70) >consensus U_AAA________UGUAUACUCACAUCAUCUGGAGCAGCAUGAGCAUGGCGUCCCGGG_GGAGUCUCCGGUGGCGCC___CACCAUAUCCACGAUGCCCUUGUCGUGCUGCUCGGU ................................(((((((((((.((.((((((.(((.........)))((((...............))))))))))..)))))))))))))... (-28.11 = -28.94 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:53 2006