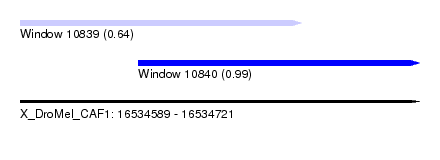
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,534,589 – 16,534,721 |
| Length | 132 |
| Max. P | 0.991171 |

| Location | 16,534,589 – 16,534,682 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -18.25 |
| Energy contribution | -21.62 |
| Covariance contribution | 3.38 |
| Combinations/Pair | 0.97 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16534589 93 + 22224390 UGCUUUAUCGAUGACUCGAGUUCUGGGCUAUAAAAGCCAUGCCAUUGAGGUCAUAAAUACAAUAAAUAAGCA--------------------UGCUUAGUUAGCGAUUAUUAC ((((((((..(((((((((......((((.....))))......)))).)))))..)))........)))))--------------------((((.....))))........ ( -20.40) >DroSec_CAF1 10228 88 + 1 AACUUUAUCGAUGACACGAAUUCUGGGCUAUAAAAGCCAUGCCAUUGAGCUCAUAAA-----UAACUAAGUA--------------------UGCUUAGUUAGCGAUUAUUAC ......(((((((((.(((......((((.....))))......))).).))))...-----((((((((..--------------------..)))))))).))))...... ( -18.70) >DroSim_CAF1 11113 88 + 1 AGCUUUAUCGAUGACUCGAGUUCUGGGCUAUAAAAGCCAUGCCAUUGAGCUCAUAAA-----UAACUAAGUG--------------------UGCUUAGUUAGCGAUUAUUAC ......(((((((((((((......((((.....))))......))))).))))...-----((((((((..--------------------..)))))))).))))...... ( -24.70) >DroEre_CAF1 4041 108 + 1 AGCUUUAUCGACGUCUCGAGUUCUGGGCAAUAAAAGCCAUGCCAUUGAGGUCAUAAA-----UAAAUAAGCAUUCCACACGCAAGACGGCGGUGCUUAGUUAGUGAUUAUUAC ....((((..(((((((((......(((.......)))......)))))).).....-----....((((((((((...(....)..)).))))))))))..))))....... ( -25.10) >consensus AGCUUUAUCGAUGACUCGAGUUCUGGGCUAUAAAAGCCAUGCCAUUGAGCUCAUAAA_____UAAAUAAGCA____________________UGCUUAGUUAGCGAUUAUUAC ......(((((((((((((......((((.....))))......))))).))))........((((((((((((((((((....)))))))))))))))))).))))...... (-18.25 = -21.62 + 3.38)


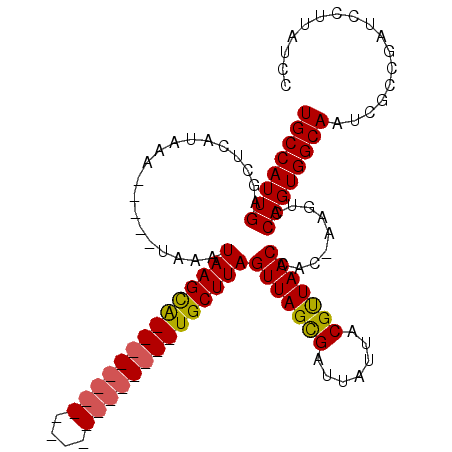
| Location | 16,534,628 – 16,534,721 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -21.79 |
| Energy contribution | -23.47 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16534628 93 + 22224390 UGCCAUUGAGGUCAUAAAUACAAUAAAUAAGCA--------------------UGCUUAGUUAGCGAUUAUUACGUUAACCAC-AAGUCCAGUGGCAAUCACCGAUCCUUAUCC ((((((((.((.(..............((((..--------------------..))))(((((((.......)))))))...-..)))))))))))................. ( -20.10) >DroSec_CAF1 10267 88 + 1 UGCCAUUGAGCUCAUAAA-----UAACUAAGUA--------------------UGCUUAGUUAGCGAUUAUUACGUUAACCAC-AAGUCCAGUGGCAAUCGCCGAUCCUUAUCC ((((((((..((......-----((((((((..--------------------..))))))))(((.......))).......-.))..))))))))................. ( -22.70) >DroSim_CAF1 11152 88 + 1 UGCCAUUGAGCUCAUAAA-----UAACUAAGUG--------------------UGCUUAGUUAGCGAUUAUUACGUUAACCAC-AAGUCCAGUGGCAAUCGCCGAUCCUUAUCC ((((((((..((......-----((((((((..--------------------..))))))))(((.......))).......-.))..))))))))................. ( -22.70) >DroEre_CAF1 4080 107 + 1 UGCCAUUGAGGUCAUAAA-----UAAAUAAGCAUUCCACACGCAAGACGGCGGUGCUUAGUUAGUGAUUAUUACGCUAACCAGU--GUCCAGUGGCAAUCGCCGAUCCUUAUCC ((((((((.((.(((...-----....((((((((((...(....)..)).))))))))(((((((.......)))))))..))--)))))))))))................. ( -31.60) >consensus UGCCAUUGAGCUCAUAAA_____UAAAUAAGCA____________________UGCUUAGUUAGCGAUUAUUACGUUAACCAC_AAGUCCAGUGGCAAUCGCCGAUCCUUAUCC ((((((((...................((((((((((((((....))))))))))))))(((((((.......))))))).........))))))))................. (-21.79 = -23.47 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:52 2006