
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,528,161 – 16,528,384 |
| Length | 223 |
| Max. P | 0.945874 |

| Location | 16,528,161 – 16,528,280 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -43.27 |
| Consensus MFE | -37.15 |
| Energy contribution | -37.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16528161 119 + 22224390 CCGCCGCCGAAGAGGCGACGC-UGCGGUAGAAACGCCGUUUUGCCGCUUGUGUUGCAAGUUGCAGGUUGCACAAUCCGGUGUUGUUUUAUGUUUUUGUUGUUACUGCUGCACGUUGCCAG .............(((((((.-((((((((.(((((((..((((.(((((((........))))))).)))..)..)))))))....((......))......))))))))))))))).. ( -41.90) >DroSec_CAF1 3926 118 + 1 NNNNNNNNNNNNNNGCGACGC-UGCGGUAGA-ACGCCGUUUUGCUGCCUGUGUUGCUAGUUGCAGGUUGCACAAUCCGGUGUUGUUUUAUGUUUUUGUUGAUACUGCUGCACGUUGCCAG ..............((((((.-(((((((((-((((((..((((.(((((((........))))))).)))..)..)))))))......((((......))))))))))))))))))... ( -42.70) >DroSim_CAF1 4193 120 + 1 CCGCCGCCGAAGAGGCGACGCGUGCUGUAGAAACGCCGUUUUGCUGCCUGUGUUGCAAGUUGCAGGUUGCACAAUCCGGUGUUGUUUUAUGUUUUUGUUGAUACUGCUGCACGUUGCCAG ..((((((.....))))..((((((.((((.(((((((..((((.(((((((........))))))).)))..)..)))))))......((((......)))))))).)))))).))... ( -43.60) >DroYak_CAF1 859 112 + 1 CCGCCGCCGAAGAGGCGACGC-UGCGGUAGAAACGCCGUCUUGUCGCCUGUGUUGCAAGUUGCAGGUUGCACAAUCCGGUGUUGU-------UGUUGUUGAUUUUACUGCAGGUUGCCAG .............((((((.(-((((((((((((((((..((((((((((((........)))))).)).))))..)))))))((-------(......))))))))))))))))))).. ( -44.90) >consensus CCGCCGCCGAAGAGGCGACGC_UGCGGUAGAAACGCCGUUUUGCCGCCUGUGUUGCAAGUUGCAGGUUGCACAAUCCGGUGUUGUUUUAUGUUUUUGUUGAUACUGCUGCACGUUGCCAG .............(((((((..((((((((.(((((((..((((.(((((((........))))))).)))..)..)))))))....................))))))))))))))).. (-37.15 = -37.40 + 0.25)



| Location | 16,528,240 – 16,528,344 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.85 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16528240 104 - 22224390 U------UAU----------AAAUUAGUUGCCCAUUUUAUAUUAACUUUUAGUGUCAAUUUAAUGUGGCAGCACUAGCAGCUGGCAACGUGCAGCAGUAACAACAAAAACAUAAAACAAC .------...----------......((((((((((..((....((.....))....))..)))).))))))(((.((.((((....)).)).)))))...................... ( -20.90) >DroSec_CAF1 4004 103 - 1 U------AAU----------AAUUUAGUUGCCCAUUUUAUAUUAACU-UUAGUGCCAAUUUAAUGUGGCAGCACUAGCAGCUGGCAACGUGCAGCAGUAUCAACAAAAACAUAAAACAAC .------...----------......((((((((((..((....((.-...))....))..)))).))))))(((.((.((((....)).)).)))))...................... ( -20.90) >DroSim_CAF1 4273 103 - 1 U------UAU----------UAUUUAGUUGCCCAUUUUAUAUUAACU-UUAGUGUCAAUUUAAUGUGGCAGCACUAGCAGCUGGCAACGUGCAGCAGUAUCAACAAAAACAUAAAACAAC .------...----------......((((((((((....(((.((.-.....)).)))..)))).))))))(((.((.((((....)).)).)))))...................... ( -21.30) >DroYak_CAF1 938 112 - 1 UAUAAUUUAUUAAAUACACCAAUUCAAUUACCCAUUAAAUAUUAAAU-UUAGUGUCAAUUUAAUGUGGCAACAGUAGCAGCUGGCAACCUGCAGUAAAAUCAACAACA-------ACAAC .....((((((...(((...............((((((((.....))-))))))...........((....)))))((((..(....)))))))))))..........-------..... ( -15.00) >consensus U______UAU__________AAUUUAGUUGCCCAUUUUAUAUUAACU_UUAGUGUCAAUUUAAUGUGGCAGCACUAGCAGCUGGCAACGUGCAGCAGUAUCAACAAAAACAUAAAACAAC ..........................((((((((((..((((((.....))))))......)))).))))))....((.((((....)).)).))......................... (-14.23 = -14.85 + 0.62)


| Location | 16,528,280 – 16,528,384 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 85.76 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -14.51 |
| Energy contribution | -14.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
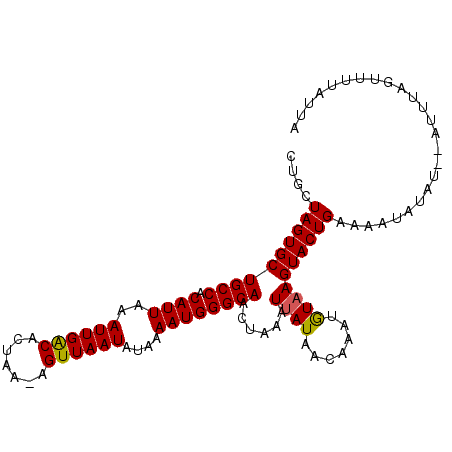
>X_DroMel_CAF1 16528280 104 + 22224390 CUGCUAGUGCUGCCACAUUAAAUUGACACUAAAAGUUAAUAUAAAAUGGGCAACUAAUUUAUAACAUAGGUAAGUACUGAAAAUAUAUAUAUUUAGUUUUAUUA ....((((((((((........(((((.......)))))((((((.(((....))).)))))).....))).)))))))......................... ( -19.80) >DroSec_CAF1 4044 101 + 1 CUGCUAGUGCUGCCACAUUAAAUUGGCACUAA-AGUUAAUAUAAAAUGGGCAACUAAAUUAUUACAAAUGUAAGUACUGAAAAUCUAC--AUUUAGUUUUAUUA ....((((((((((.((((..((((((.....-.))))))....)))))))).........((((....)))))))))).........--.............. ( -17.70) >DroSim_CAF1 4313 100 + 1 CUGCUAGUGCUGCCACAUUAAAUUGACACUAA-AGUUAAUAUAAAAUGGGCAACUAAAUAAUAAGAAGUAUAAGUACUGAA-GUAUAU--AAGUACUGUAAGUA ..(((.((((((((.((((..((((((.....-.))))))....)))))))..((........)).))))).)))(((..(-((((..--..)))))...))). ( -20.10) >consensus CUGCUAGUGCUGCCACAUUAAAUUGACACUAA_AGUUAAUAUAAAAUGGGCAACUAAAUUAUAACAAAUGUAAGUACUGAAAAUAUAU__AUUUAGUUUUAUUA ....((((((((((.((((..((((((.......))))))....))))))))......((((.......))))))))))......................... (-14.51 = -14.40 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:44 2006