
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,512,119 – 16,512,284 |
| Length | 165 |
| Max. P | 0.999990 |

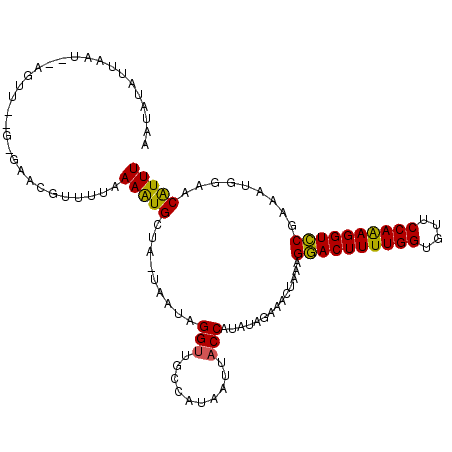
| Location | 16,512,119 – 16,512,231 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.21 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -17.34 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.64 |
| SVM decision value | 5.56 |
| SVM RNA-class probability | 0.999990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16512119 112 + 22224390 AAUAAAUUAUUGUAGUUGAGAGAACGUUUUAAAAUGCUA-UAAUAGGUUGCCAUAAUUACCAUAUAGAAACUUAAGAACUUUUGGUGUUCCAAAGGUUCGAAAUGGAACAUUU ......(((((((((((((((.....)))))....))))-))))))(((.((((..........(((....))).((((((((((....))))))))))...))))))).... ( -23.40) >DroSec_CAF1 8782 108 + 1 AAUAUAUUAC-----UUCUGGGAACGUUUUAAAAUGCUAAUGAUGGGUUGCCAUAAUUACCAUAUAGAAACUAAAGGACUUUUGGUGUUCCAAAGGUCCGAACUGGAACGUUU ..........-----......((((((((((..(((.((((.((((....)))).)))).)))............((((((((((....))))))))))....)))))))))) ( -30.50) >DroEre_CAF1 8766 95 + 1 AAUAUAUCAAU--AG-------------UUCAAGUGGUA-UAAUAGGU-ACCAUAAUUCCCAUAUA-AAACUAUAGGACUUUUGGUAUCCCAGAGGUCCAAAAAUGAACAUUU ...........--.(-------------((((.((((((-(.....))-)))))............-........((((((((((....)))))))))).....))))).... ( -27.90) >consensus AAUAUAUUAAU__AGUU__G_GAACGUUUUAAAAUGCUA_UAAUAGGUUGCCAUAAUUACCAUAUAGAAACUAAAGGACUUUUGGUGUUCCAAAGGUCCGAAAUGGAACAUUU ...............................(((((.........(((..........)))..............((((((((((....)))))))))).........))))) (-17.34 = -16.57 + -0.77)

| Location | 16,512,119 – 16,512,231 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.21 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -11.88 |
| Energy contribution | -12.43 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16512119 112 - 22224390 AAAUGUUCCAUUUCGAACCUUUGGAACACCAAAAGUUCUUAAGUUUCUAUAUGGUAAUUAUGGCAACCUAUUA-UAGCAUUUUAAAACGUUCUCUCAACUACAAUAAUUUAUU .((((((.......((((.(((((....))))).))))(((((...(((((.(((..........)))...))-)))...)))))))))))...................... ( -16.00) >DroSec_CAF1 8782 108 - 1 AAACGUUCCAGUUCGGACCUUUGGAACACCAAAAGUCCUUUAGUUUCUAUAUGGUAAUUAUGGCAACCCAUCAUUAGCAUUUUAAAACGUUCCCAGAA-----GUAAUAUAUU .........(.(((((((.(((((....))))).))))....((((..(.(((.((((.((((....)))).)))).))).)..)))).......)))-----.)........ ( -22.80) >DroEre_CAF1 8766 95 - 1 AAAUGUUCAUUUUUGGACCUCUGGGAUACCAAAAGUCCUAUAGUUU-UAUAUGGGAAUUAUGGU-ACCUAUUA-UACCACUUGAA-------------CU--AUUGAUAUAUU ....(((((....(((.....((((.(((((.((.(((((((....-..))))))).)).))))-)))))...-..)))..))))-------------).--........... ( -25.40) >consensus AAAUGUUCCAUUUCGGACCUUUGGAACACCAAAAGUCCUUUAGUUUCUAUAUGGUAAUUAUGGCAACCUAUUA_UAGCAUUUUAAAACGUUC_C__AACU__AAUAAUAUAUU ..............((((.(((((....))))).))))....(((.(((((.......))))).))).............................................. (-11.88 = -12.43 + 0.56)


| Location | 16,512,156 – 16,512,260 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -16.12 |
| Energy contribution | -15.57 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16512156 104 + 22224390 UA-UAAUAGGUUGCCAUAAUUACCAUAUAGAAACUUAAGAACUUUUGGUGUUCCAAAGGUUCGAAAUGGAACAUUUUUAUUU-----------AACUAAUAUCUUGAAAGCGAAAA ..-.......((((........................((((((((((....)))))))))).(((((((.....)))))))-----------................))))... ( -17.20) >DroSec_CAF1 8814 116 + 1 UAAUGAUGGGUUGCCAUAAUUACCAUAUAGAAACUAAAGGACUUUUGGUGUUCCAAAGGUCCGAACUGGAACGUUUUUAUAUUUUUAUCAUGGAACUUAUAUCUCGAAAGCGAAAA .....(((((((.((((.((....((((((((((....((((((((((....))))))))))(........)))))))))))....)).))))))))))).((.(....).))... ( -33.20) >DroEre_CAF1 8788 100 + 1 UA-UAAUAGGU-ACCAUAAUUCCCAUAUA-AAACUAUAGGACUUUUGGUAUCCCAGAGGUCCAAAAAUGAACAUUUUAAUUC-----------AACUU-UAUCUCGAAACCGAA-A ..-.....(((-.................-........((((((((((....)))))))))).....((((........)))-----------)....-.........)))...-. ( -19.00) >consensus UA_UAAUAGGUUGCCAUAAUUACCAUAUAGAAACUAAAGGACUUUUGGUGUUCCAAAGGUCCGAAAUGGAACAUUUUUAUUU___________AACUUAUAUCUCGAAAGCGAAAA ........(((..........)))..............((((((((((....)))))))))).........................................(((....)))... (-16.12 = -15.57 + -0.55)

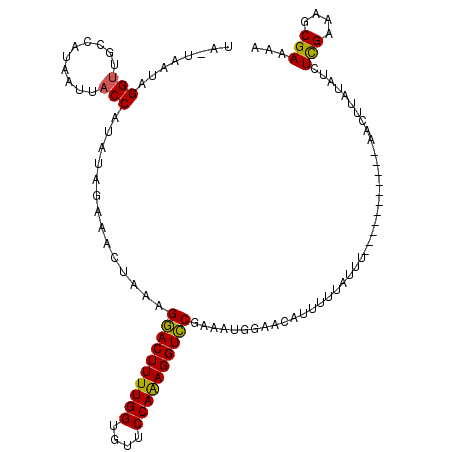
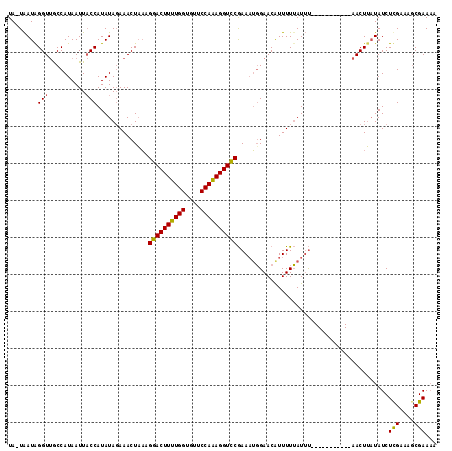
| Location | 16,512,191 – 16,512,284 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.37 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -15.82 |
| Energy contribution | -14.93 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.67 |
| SVM decision value | 5.00 |
| SVM RNA-class probability | 0.999968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16512191 93 + 22224390 AAGAACUUUUGGUGUUCCAAAGGUUCGAAAUGGAACAUUUUUAUUU-----------AACUAAUAUCUUGAAAGCGAAAAAAGCAAAAGAUUUUU-GA---------------UAAGUGU ..((((((((((....))))))))))((((((...))))))(((((-----------(.(....(((((....((.......))..)))))....-).---------------)))))). ( -20.10) >DroSec_CAF1 8850 105 + 1 AAGGACUUUUGGUGUUCCAAAGGUCCGAACUGGAACGUUUUUAUAUUUUUAUCAUGGAACUUAUAUCUCGAAAGCGAAAAAAGUAAAAGAUUUUUGGA---------------UAAGUGU ..((((((((((....))))))))))...(..(((.((((((.(((((((.((..(((.......)))(....).)).))))))))))))))))..).---------------....... ( -24.80) >DroEre_CAF1 8821 106 + 1 UAGGACUUUUGGUAUCCCAGAGGUCCAAAAAUGAACAUUUUAAUUC-----------AACUU-UAUCUCGAAACCGAA-AAUGCAAUAGAUUUUU-GAUUUUUGUUUAGUUUCUGAGUUU ..((((((((((....)))))))))).....((((........)))-----------)....-...((((((((((((-(((.(((.......))-))))))))....))))).)))... ( -25.60) >consensus AAGGACUUUUGGUGUUCCAAAGGUCCGAAAUGGAACAUUUUUAUUU___________AACUUAUAUCUCGAAAGCGAAAAAAGCAAAAGAUUUUU_GA_______________UAAGUGU ..((((((((((....)))))))))).........................................(((....)))........................................... (-15.82 = -14.93 + -0.88)

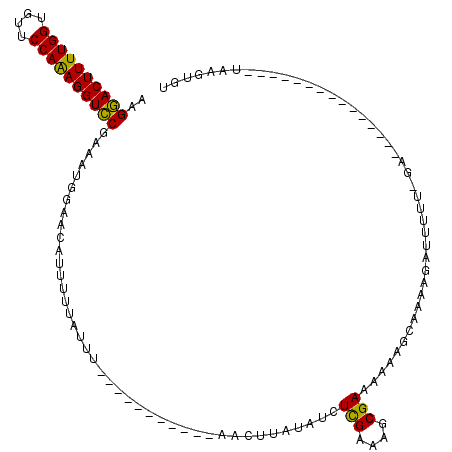
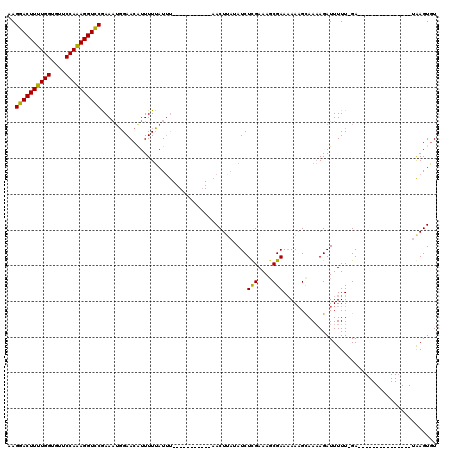
| Location | 16,512,191 – 16,512,284 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.37 |
| Mean single sequence MFE | -16.03 |
| Consensus MFE | -10.43 |
| Energy contribution | -10.77 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16512191 93 - 22224390 ACACUUA---------------UC-AAAAAUCUUUUGCUUUUUUCGCUUUCAAGAUAUUAGUU-----------AAAUAAAAAUGUUCCAUUUCGAACCUUUGGAACACCAAAAGUUCUU ..((((.---------------..-....(((((..((.......))....))))).......-----------.........(((((((...........)))))))....)))).... ( -11.70) >DroSec_CAF1 8850 105 - 1 ACACUUA---------------UCCAAAAAUCUUUUACUUUUUUCGCUUUCGAGAUAUAAGUUCCAUGAUAAAAAUAUAAAAACGUUCCAGUUCGGACCUUUGGAACACCAAAAGUCCUU .......---------------.......(((....((((.(((((....)))))...)))).....)))........................((((.(((((....))))).)))).. ( -16.90) >DroEre_CAF1 8821 106 - 1 AAACUCAGAAACUAAACAAAAAUC-AAAAAUCUAUUGCAUU-UUCGGUUUCGAGAUA-AAGUU-----------GAAUUAAAAUGUUCAUUUUUGGACCUCUGGGAUACCAAAAGUCCUA ...(((.((((((.....((((((-((.......))).)))-)).)))))))))..(-(((.(-----------((((......))))).))))((((...(((....)))...)))).. ( -19.50) >consensus ACACUUA_______________UC_AAAAAUCUUUUGCUUUUUUCGCUUUCGAGAUAUAAGUU___________AAAUAAAAAUGUUCCAUUUCGGACCUUUGGAACACCAAAAGUCCUU ....................................((((...((........))...))))................................((((.(((((....))))).)))).. (-10.43 = -10.77 + 0.34)

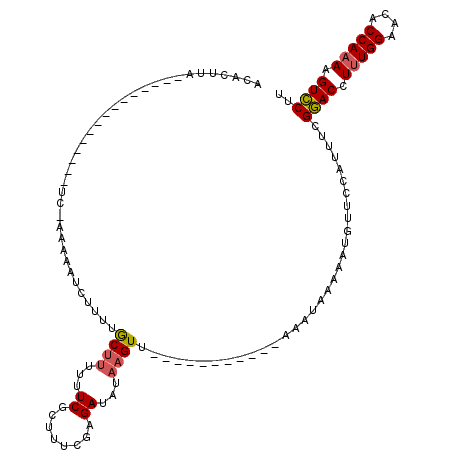

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:35 2006