| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,508,429 – 16,508,527 |
| Length | 98 |
| Max. P | 0.963299 |

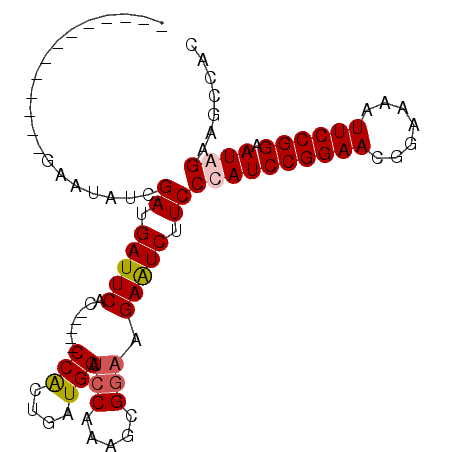
| Location | 16,508,429 – 16,508,527 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16508429 98 + 22224390 GUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCAGUGGGG---GUGAAUCAUCGAUACUCGAUAAUCGAUUCU .......((((.(((((((.....))))))))))).......((((((((.((....))..)))))))---).(((((((((.....))))....))))). ( -31.70) >DroSec_CAF1 5179 101 + 1 GUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCUGUGGAGUGUGUGAAUCAUCGAUACUCGAUACUCUAUUCU ((((...(((.((((((((.....))))))))..(((((...)))))....)))..))))..((((((((((.(.(((...))).).).)))))))))... ( -29.50) >DroEre_CAF1 5171 83 + 1 GUGAGUUCUAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGAUUCUUCCGCUUUGGAGCCAUCAGCGG-----GUGAAUCAUCGGUAUUC------------- .(((.((((((.(((((((.....))))))))))))).(((((.((((((.((....))..)))))-----).))))).)))......------------- ( -31.10) >DroYak_CAF1 5035 83 + 1 GUGGCUUCUAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGAUUCUUCCGCUUUGCCUCCAUCAGUGG-----GUGAAUCAUCGAUAUUC------------- .....((((((.(((((((.....))))))))))))).(((((.((((((.((....))..)))))-----).)))))..........------------- ( -26.60) >consensus GUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCAGUGG_____GUGAAUCAUCGAUACUC_____________ .......((((.(((((((.....)))))))))))((.(((((((((((..((....))...)))))).....))))).)).................... (-22.70 = -23.70 + 1.00)

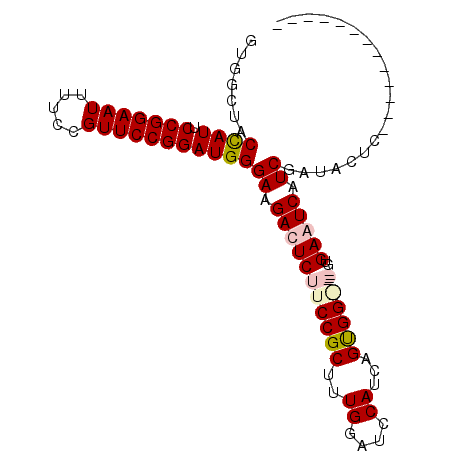

| Location | 16,508,429 – 16,508,527 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16508429 98 - 22224390 AGAAUCGAUUAUCGAGUAUCGAUGAUUCAC---CCCCACUGAUGGAUCCAAAGCGGAAGAGUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUGGUAGCCAC ......((((((((.....))))))))...---.........(((..(((..(.(((((...)))))).(((((((.......))))))).)))...))). ( -27.50) >DroSec_CAF1 5179 101 - 1 AGAAUAGAGUAUCGAGUAUCGAUGAUUCACACACUCCACAGAUGGAUCCAAAGCGGAAGAGUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUGGUAGCCAC ......((((((((.....)))).))))..............(((..(((..(.(((((...)))))).(((((((.......))))))).)))...))). ( -26.70) >DroEre_CAF1 5171 83 - 1 -------------GAAUACCGAUGAUUCAC-----CCGCUGAUGGCUCCAAAGCGGAAGAAUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUAGAACUCAC -------------.......((.(((((..-----(((((..((....)).)))))..))))).))...(((((((.......)))))))........... ( -25.70) >DroYak_CAF1 5035 83 - 1 -------------GAAUAUCGAUGAUUCAC-----CCACUGAUGGAGGCAAAGCGGAAGAAUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUAGAAGCCAC -------------...(((((.((......-----.)).)))))..(((...(.(((((...)))))).(((((((.......)))))))......))).. ( -22.30) >consensus _____________GAAUAUCGAUGAUUCAC_____CCACUGAUGGAUCCAAAGCGGAAGAAUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUAGAAGCCAC ....................((.(((((.......(((....))).(((.....))).))))).))((((((((((.......)))))).))))....... (-17.19 = -17.75 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:30 2006