| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,507,198 – 16,507,293 |
| Length | 95 |
| Max. P | 0.653656 |

| Location | 16,507,198 – 16,507,293 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -17.21 |
| Consensus MFE | -11.21 |
| Energy contribution | -12.10 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
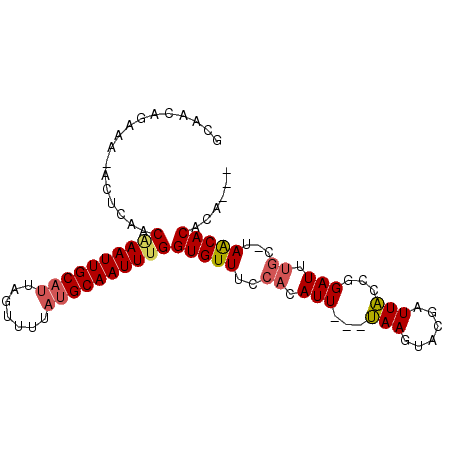
>X_DroMel_CAF1 16507198 95 + 22224390 GCAACAGAAAGACUCAACAAAUUGCAUUAGUUUUAUGCAAUUUGGUGUUUCCACAUUUAUUAAGUACGAUUACCGGAUUUGCCUAACACACACAC .................((((((((((.......))))))))))(((((..((.((((..(((......)))..)))).))...)))))...... ( -16.80) >DroSec_CAF1 4007 87 + 1 GCAACAGAAAAACUCAACAAAUUGCAUUAGUUUUACGCAAUUCGGUGUUUCCACAUU---UAAGUACGAUUACCGGAUUUGCUUAACACA----- ((((.................))))....((.(((.(((((((((((.((..((...---...))..)).))))))).)))).))).)).----- ( -15.13) >DroEre_CAF1 4042 81 + 1 GCAACAGAAA---ACAACGAAUUGCAGUACUUUUAUGCAAUUUGGUGUUUCCACAUU---GAAGUGCUUUUUCUUGAUU-----AGCACACA--- ......(.((---(((.(((((((((.........))))))))).))))).).....---...(((((..((...))..-----)))))...--- ( -19.70) >consensus GCAACAGAAA_ACUCAACAAAUUGCAUUAGUUUUAUGCAAUUUGGUGUUUCCACAUU___UAAGUACGAUUACCGGAUUUGC_UAACACACA___ .................((((((((((.......))))))))))(((((..((.(((...(((......)))...))).))...)))))...... (-11.21 = -12.10 + 0.89)

| Location | 16,507,198 – 16,507,293 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -13.15 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16507198 95 - 22224390 GUGUGUGUGUUAGGCAAAUCCGGUAAUCGUACUUAAUAAAUGUGGAAACACCAAAUUGCAUAAAACUAAUGCAAUUUGUUGAGUCUUUCUGUUGC ((.((.....)).)).......(((((.(.((((((.....(((....)))((((((((((.......))))))))))))))))....).))))) ( -20.70) >DroSec_CAF1 4007 87 - 1 -----UGUGUUAAGCAAAUCCGGUAAUCGUACUUA---AAUGUGGAAACACCGAAUUGCGUAAAACUAAUGCAAUUUGUUGAGUUUUUCUGUUGC -----........((((...(((.......(((((---(..(((....)))((((((((((.......))))))))))))))))....))))))) ( -20.70) >DroEre_CAF1 4042 81 - 1 ---UGUGUGCU-----AAUCAAGAAAAAGCACUUC---AAUGUGGAAACACCAAAUUGCAUAAAAGUACUGCAAUUCGUUGU---UUUCUGUUGC ---...(((((-----...........)))))..(---((((.(((((((.(.(((((((.........))))))).).)))---))))))))). ( -18.90) >consensus ___UGUGUGUUA_GCAAAUCCGGUAAUCGUACUUA___AAUGUGGAAACACCAAAUUGCAUAAAACUAAUGCAAUUUGUUGAGU_UUUCUGUUGC ............................(((..........(((....)))((((((((((.......))))))))))..............))) (-13.15 = -13.27 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:28 2006