
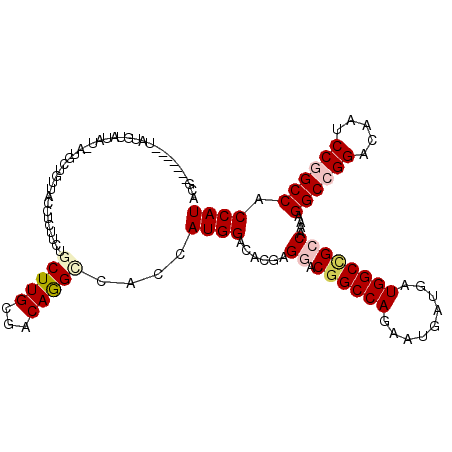
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,503,222 – 16,503,335 |
| Length | 113 |
| Max. P | 0.696897 |

| Location | 16,503,222 – 16,503,335 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.63 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16503222 113 + 22224390 AGC--AUAUGUAUGUAUGU-AUGCUGUUACCUCUUCUGCUUGCGACAAGAUACCAUGGACACGAGGACGGCCAAAAUGAUGAUGGCCGCCAAAAGGCCGGAUAAUCCGGCCACCAU (((--((((((....))))-))))).............((((...)))).....((((......((.((((((.........))))))))....((((((.....)))))).)))) ( -38.90) >DroPse_CAF1 272 113 + 1 ACUGCAU---UAUGUAUACGAAACUCUUACCUCUUCUGCUUGCGGCAGGCCACCAUGGAGGCGAGCACGGCCAGAAUGAUAAUGGCUGCCAGUAGGCCGGACAGACCCGCCACCAU ((((...---...............((..((((((((((.....))))).......)))))..))..((((((.........)))))).)))).(((.((.....)).)))..... ( -30.71) >DroSec_CAF1 237 108 + 1 -------AUAUAUGUAUGU-AUGCUGUUACCUCUUCUGCUUGCGACAAGAUAUCAUGGACACAAGGACGGCCAGAAUGAUGAUGGCAGCCAAAAGGCCGGACAAUCCGGCCACCAU -------.....(((.(((-(.((.............)).))))))).......((((......((...((((.........))))..))....((((((.....)))))).)))) ( -30.82) >DroEre_CAF1 256 103 + 1 AGG------------GCAU-AUGCUGUUACCUCUUCUGCUUGCGACAGGCCACCAUGGACACGAGGACGGCCAGAAUGAUGAUGGCCGCCAAAAGGCCGGAUAAUCCGGCCACCAU ..(------------((..-.((.(((...(......)...))).)).)))...((((......((.((((((.........))))))))....((((((.....)))))).)))) ( -38.10) >DroYak_CAF1 231 95 + 1 AUG---------------------UGUUACCUCUUCUGCUUGCGACAGGUCACCAUGGACACGAGGACGGCCAGAAUGAUGAUGGCCGCCAAAAGGCCGGACAAUCCGGCCACCAU .((---------------------((((......(((((....).))))........)))))).((.((((((.........))))))))....((((((.....))))))..... ( -35.54) >DroPer_CAF1 982 113 + 1 ACUGCAU---UAUGUAUACGAAACUCUUACCUCUUCUGCUUGCGACAGGCCACCAUGGAGGCGAGCACGGCCAGAAUGAUAAUGGCUGCCAGUAGGCCGGACAGACCCGCCACCAU ((((...---...............((..(((((...(((((...)))))......)))))..))..((((((.........)))))).)))).(((.((.....)).)))..... ( -29.60) >consensus ACG_______UAUGUAUAU_AUGCUGUUACCUCUUCUGCUUGCGACAGGCCACCAUGGACACGAGGACGGCCAGAAUGAUGAUGGCCGCCAAAAGGCCGGACAAUCCGGCCACCAU .....................................(((((...)))))....((((......((.((((((.........))))))))....((((((.....)))))).)))) (-25.94 = -26.63 + 0.69)


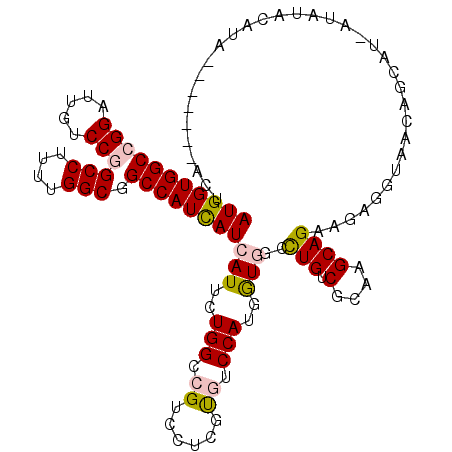
| Location | 16,503,222 – 16,503,335 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.23 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16503222 113 - 22224390 AUGGUGGCCGGAUUAUCCGGCCUUUUGGCGGCCAUCAUCAUUUUGGCCGUCCUCGUGUCCAUGGUAUCUUGUCGCAAGCAGAAGAGGUAACAGCAU-ACAUACAUACAUAU--GCU ((((.((((((.....))))))....((((((((.........)))))))).......))))(.((((((.((.......)).)))))).)(((((-(..........)))--))) ( -38.80) >DroPse_CAF1 272 113 - 1 AUGGUGGCGGGUCUGUCCGGCCUACUGGCAGCCAUUAUCAUUCUGGCCGUGCUCGCCUCCAUGGUGGCCUGCCGCAAGCAGAAGAGGUAAGAGUUUCGUAUACAUA---AUGCAGU (((((((((((....))).(((....))).)))))))).(((((.(((..((.((((.....))))))(((.(....))))....))).)))))...((((.....---))))... ( -37.50) >DroSec_CAF1 237 108 - 1 AUGGUGGCCGGAUUGUCCGGCCUUUUGGCUGCCAUCAUCAUUCUGGCCGUCCUUGUGUCCAUGAUAUCUUGUCGCAAGCAGAAGAGGUAACAGCAU-ACAUACAUAUAU------- ..((((((((((.((..(((((....))))).......)).)))))))).)).(((((...((.((((((.((.......)).)))))).))..))-))).........------- ( -33.80) >DroEre_CAF1 256 103 - 1 AUGGUGGCCGGAUUAUCCGGCCUUUUGGCGGCCAUCAUCAUUCUGGCCGUCCUCGUGUCCAUGGUGGCCUGUCGCAAGCAGAAGAGGUAACAGCAU-AUGC------------CCU .((((((((((.....)).(((....)))))))))))((.(((((((((.((..........))))))).(.(....))))))))((((.......-.)))------------).. ( -37.60) >DroYak_CAF1 231 95 - 1 AUGGUGGCCGGAUUGUCCGGCCUUUUGGCGGCCAUCAUCAUUCUGGCCGUCCUCGUGUCCAUGGUGACCUGUCGCAAGCAGAAGAGGUAACA---------------------CAU .....((((((.....))))))....((((((((.........))))))))...((((((..(....)(((.(....))))....))..)))---------------------).. ( -37.80) >DroPer_CAF1 982 113 - 1 AUGGUGGCGGGUCUGUCCGGCCUACUGGCAGCCAUUAUCAUUCUGGCCGUGCUCGCCUCCAUGGUGGCCUGUCGCAAGCAGAAGAGGUAAGAGUUUCGUAUACAUA---AUGCAGU (((((((((((....))).(((....))).)))))))).(((((.(((..((.((((.....))))))(((.(....))))....))).)))))...((((.....---))))... ( -37.50) >consensus AUGGUGGCCGGAUUGUCCGGCCUUUUGGCGGCCAUCAUCAUUCUGGCCGUCCUCGUGUCCAUGGUGGCCUGUCGCAAGCAGAAGAGGUAACAGCAU_AUAUACAUA_______ACU (((((((((((.....)))(((....))).))))))))(((..(((.((......)).)))..)))..(((.(....))))................................... (-25.04 = -25.23 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:27 2006