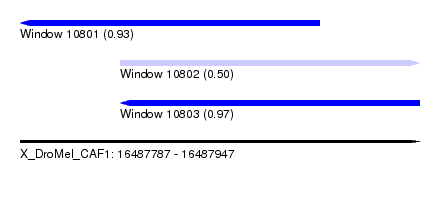
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,487,787 – 16,487,947 |
| Length | 160 |
| Max. P | 0.974351 |

| Location | 16,487,787 – 16,487,907 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -46.37 |
| Consensus MFE | -38.00 |
| Energy contribution | -39.04 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16487787 120 - 22224390 CUGGCACACCAGCGCCGUGAUCUUCAUCAUGGUGCUGGUCUUUGUGGCCCACUACUUCCCCAUUUCGUGGACGAACCAGAUCAAGUUCAUGUAUCGCCGCCGUUUUCCGCUAAAGAGGCG ..(((..((((((((((((((....))))))))))))))......(((((((..............))))..((((........)))).......))))))......((((.....)))) ( -43.34) >DroSec_CAF1 25962 120 - 1 CUGGUACACGAGCGCCGUCGCCUUGAUCAUGGUGCUGGUCUUUGCGGCCCACUACUUCCCCAGUUCGUGGGCGAACCAGAUCAAGACCAUGUACCGCCGCCGCUUUCCGCCAAAUAGGCG ..((((((.(.(((....)))(((((((.((((((........)).((((((.(((.....)))..))))))..)))))))))))..).))))))((....))....((((.....)))) ( -43.70) >DroSim_CAF1 23071 120 - 1 CUGGUACACCAGCGCCGUGGCCUUCAUCAUGGUGCUGGUCUUUGCGGCCCACUACUUCCCCAGUUCGUGGGCGAACCAGAUCAAGACCAUGUACCGCCGCCGCUUUCCGCCAAAGAGGCG ..((((((..((((((((((......))))))))))((((((....((((((.(((.....)))..))))))((......)))))))).))))))..((((.((((.....)))).)))) ( -50.20) >DroEre_CAF1 16965 120 - 1 CUGGCACACCAGCGCCCUGGUCUUUAUCAUGGUUCUGCUCUUUGUGGCCUACUACUUUCCCAGGUCGUGGGCCCGCCAGGUCAAGAUCAUGUACCGUCGCCGCUUUCCGCCAAAGAGGCG ..((((((((((....)))))......((((((..((..(((.(((((((((.(((......))).))))).)))).))).))..))))))....)).)))......((((.....)))) ( -41.60) >DroYak_CAF1 18017 120 - 1 CUGGCACACCAGCGCCCUGGUCUUCAUCACGGUGCUGGUCUUUGUGGCCCACUACUUUCCCAGGUCGUGGGCGCGCCAGGCGAAGAUCAUGUACCGCCGCCGCUUUCCGCCAAAGAGGCG .((((.((((((((((.((((....)))).))))))))).......((((((.(((......))).))))))).))))((((............))))(((.((((.....)))).))). ( -53.00) >consensus CUGGCACACCAGCGCCGUGGUCUUCAUCAUGGUGCUGGUCUUUGUGGCCCACUACUUCCCCAGUUCGUGGGCGAACCAGAUCAAGAUCAUGUACCGCCGCCGCUUUCCGCCAAAGAGGCG (((((..(((((((((((((......))))))))))))).......((((((..............))))))..)))))..................((((.((((.....)))).)))) (-38.00 = -39.04 + 1.04)


| Location | 16,487,827 – 16,487,947 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16487827 120 + 22224390 UCUGGUUCGUCCACGAAAUGGGGAAGUAGUGGGCCACAAAGACCAGCACCAUGAUGAAGAUCACGGCGCUGGUGUGCCAGCUGCUGAAGUAAGCCAUAACGGCGCCAAUGGCCUGCAGCA (((..((((....))))..)))...((.(..(((((.....((((((.((.((((....)))).)).))))))(((((....(((......)))......)))))...)))))..).)). ( -46.00) >DroSec_CAF1 26002 120 + 1 UCUGGUUCGCCCACGAACUGGGGAAGUAGUGGGCCGCAAAGACCAGCACCAUGAUCAAGGCGACGGCGCUCGUGUACCAGCUGCUUAAGUAAACCAUGAAGCCGCCUAUGGCCUGCAGCA ((..(((((....)))))..))...((.(..((((((........))..........((((...(((..(((((.....(((.....)))....))))).)))))))..))))..).)). ( -41.00) >DroSim_CAF1 23111 120 + 1 UCUGGUUCGCCCACGAACUGGGGAAGUAGUGGGCCGCAAAGACCAGCACCAUGAUGAAGGCCACGGCGCUGGUGUACCAGCUGCUUAAGUAAACCAUGAAGCCGCCUAUGGCCUGCAGCA .((((((.((((((..(((.....))).))))))......)))))).....((.((.(((((((((((((((....)))))(((....))).........))))....)))))).)).)) ( -45.70) >DroEre_CAF1 17005 120 + 1 CCUGGCGGGCCCACGACCUGGGAAAGUAGUAGGCCACAAAGAGCAGAACCAUGAUAAAGACCAGGGCGCUGGUGUGCCAGCUGCUGAAGUAAACCAGGACACCGCCAAUGGCUUGCAGCA ..((((((........(((((...((((((.((((((....(((....((.((........))))..))).))).))).))))))........)))))...))))))...((.....)). ( -39.20) >DroYak_CAF1 18057 120 + 1 CCUGGCGCGCCCACGACCUGGGAAAGUAGUGGGCCACAAAGACCAGCACCGUGAUGAAGACCAGGGCGCUGGUGUGCCAGCUGCUGAAGUAAACCAGGACAACGCCCAUGGCCUGCAGCA .(((((((((((((...((.....))..)))))).......((((((.((.((........)).)).)))))))))))))..((((.((....(((((.......)).))).)).)))). ( -50.70) >consensus UCUGGUUCGCCCACGAACUGGGGAAGUAGUGGGCCACAAAGACCAGCACCAUGAUGAAGACCACGGCGCUGGUGUGCCAGCUGCUGAAGUAAACCAUGACGCCGCCAAUGGCCUGCAGCA ..((((..((((((...((.....))..)))))).....(.((((((.((.((........)).)).)))))).)))))(((((....(....).........(((...)))..))))). (-34.06 = -34.62 + 0.56)



| Location | 16,487,827 – 16,487,947 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -48.85 |
| Consensus MFE | -40.28 |
| Energy contribution | -41.56 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16487827 120 - 22224390 UGCUGCAGGCCAUUGGCGCCGUUAUGGCUUACUUCAGCAGCUGGCACACCAGCGCCGUGAUCUUCAUCAUGGUGCUGGUCUUUGUGGCCCACUACUUCCCCAUUUCGUGGACGAACCAGA (((((.(((((...)))(((.....)))...)).))))).(((((((((((((((((((((....))))))))))))))....)))..((((..............)))).....)))). ( -48.14) >DroSec_CAF1 26002 120 - 1 UGCUGCAGGCCAUAGGCGGCUUCAUGGUUUACUUAAGCAGCUGGUACACGAGCGCCGUCGCCUUGAUCAUGGUGCUGGUCUUUGCGGCCCACUACUUCCCCAGUUCGUGGGCGAACCAGA .((((((((((......)))))...((....))...)))))((((.(.((((.((((.((((........)))).)))).)))).)((((((.(((.....)))..))))))..)))).. ( -42.80) >DroSim_CAF1 23111 120 - 1 UGCUGCAGGCCAUAGGCGGCUUCAUGGUUUACUUAAGCAGCUGGUACACCAGCGCCGUGGCCUUCAUCAUGGUGCUGGUCUUUGCGGCCCACUACUUCCCCAGUUCGUGGGCGAACCAGA .((((((((((......)))))...((....))...)))))((((.((((((((((((((......))))))))))))).......((((((.(((.....)))..))))))).)))).. ( -51.00) >DroEre_CAF1 17005 120 - 1 UGCUGCAAGCCAUUGGCGGUGUCCUGGUUUACUUCAGCAGCUGGCACACCAGCGCCCUGGUCUUUAUCAUGGUUCUGCUCUUUGUGGCCUACUACUUUCCCAGGUCGUGGGCCCGCCAGG .(((((.........)))))..((((((.......(((((((((....)))))(((.((((....)))).)))..))))....(.(((((((.(((......))).)))))))))))))) ( -44.00) >DroYak_CAF1 18057 120 - 1 UGCUGCAGGCCAUGGGCGUUGUCCUGGUUUACUUCAGCAGCUGGCACACCAGCGCCCUGGUCUUCAUCACGGUGCUGGUCUUUGUGGCCCACUACUUUCCCAGGUCGUGGGCGCGCCAGG (((((.((((((.((((...))))))))...)).))))).(((((.((((((((((.((((....)))).))))))))).......((((((.(((......))).))))))).))))). ( -58.30) >consensus UGCUGCAGGCCAUAGGCGGCGUCAUGGUUUACUUCAGCAGCUGGCACACCAGCGCCGUGGUCUUCAUCAUGGUGCUGGUCUUUGUGGCCCACUACUUCCCCAGUUCGUGGGCGAACCAGA (((((.(((((((.((.....)))))))))....))))).(((((..(((((((((((((......))))))))))))).......((((((..............))))))..))))). (-40.28 = -41.56 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:18 2006