
| Sequence ID | X_DroMel_CAF1 |
|---|---|
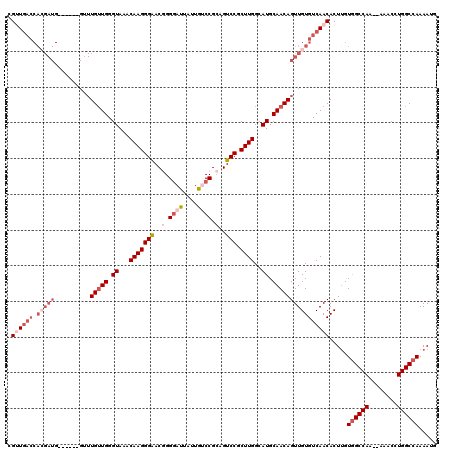
| Location | 16,470,752 – 16,470,944 |
| Length | 192 |
| Max. P | 0.860583 |

| Location | 16,470,752 – 16,470,864 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.91 |
| Mean single sequence MFE | -37.94 |
| Consensus MFE | -26.76 |
| Energy contribution | -29.96 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16470752 112 + 22224390 CGUUGACCACGAUG------GAUUGUUGGGUAAACAAGGGAACGGGGAUUAUUGUCCGCAGUCCGCUUGGCAUGCAACAGUUGUGUCAACACUUGUGGCCAA--AAACCUGGCCAAAAUG .((((((.(((((.------...(((((.((...(((((((..(.((((....)))).)..))).))))..)).)))))))))))))))).....((((((.--.....))))))..... ( -42.60) >DroSec_CAF1 8764 112 + 1 CGCUGACCACGAUG------GUUUGUUGGGUAAACAAGGGAACGGGGAUUAUUGUCCGCAGUCCUCUUGGCAUGCAACAGUUGUGUCAACACUGGUGGCCAA--AAACCUGGCCAAAAUG .(.((((.(((((.------...(((((.((...(((((((..(.((((....)))).)..))).))))..)).)))))))))))))).).....((((((.--.....))))))..... ( -38.40) >DroSim_CAF1 6027 112 + 1 CGCUGACCACGAUG------GUUUGUUGGGUAAACAAGGGAACGGGGAUUAUUGUCCGCAGUCCGCUUGGCAUGCAACAGUUGUGUCAACACUUGUGGCCAA--AAACCUGGCCAAAAUG .(.((((.(((((.------...(((((.((...(((((((..(.((((....)))).)..))).))))..)).)))))))))))))).).....((((((.--.....))))))..... ( -38.40) >DroEre_CAF1 101 96 + 1 GGUU-------------------UG-UGGGUAAACAAGGGGACAGGAACUAUUGGCCG-AGUCCGCUUGGCAUGCAACAGUUGUGUCAACACUUGUAGCCAA--AAACCUGGCCAAAAA- ((((-------------------(.-..(((..(((((..((((.((.((.(((((((-((....))))))...))).)))).))))....))))).)))..--)))))..........- ( -31.20) >DroYak_CAF1 763 118 + 1 GGUUGACCAGGAUGCAGGAUGUUUGUUGGGUAAACAAGGGGACAGGGAUUAUUGGCCG-AGUCCGCUUGGCAUGCAACAGUUGUGUCAACACUUGUGGCCAAAAAAACCUGGCCAAAAA- .(((((((((...((.....))((((((.((...((((.((((..((........)).-.)))).))))..)).))))))))).)))))).....((((((........))))))....- ( -39.10) >consensus CGUUGACCACGAUG______GUUUGUUGGGUAAACAAGGGAACGGGGAUUAUUGUCCGCAGUCCGCUUGGCAUGCAACAGUUGUGUCAACACUUGUGGCCAA__AAACCUGGCCAAAAUG .((((((.(((((..........(((((.((...(((((((..(.((((....)))).)..))).))))..)).)))))))))))))))).....((((((........))))))..... (-26.76 = -29.96 + 3.20)


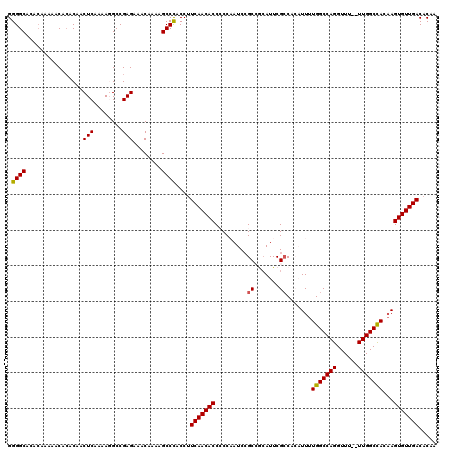
| Location | 16,470,826 – 16,470,944 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.22 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -21.29 |
| Energy contribution | -21.09 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16470826 118 - 22224390 GGGGCACACAAAAACCAACAACUCAAAAGGCCGAGAAACAAAAGCCCACCUUCAACACCACCAAUCCGCCGCAUUCGACACAUUUUGGCCAGGUUU--UUGGCCACAAGUGUUGACACAA (.(((................(((........)))......(((.....)))...............))).)..(((((((.((.(((((((....--))))))).)))))))))..... ( -26.00) >DroSec_CAF1 8838 118 - 1 GGGGCACACAAAAACACACAACUCAAAAGGCCGAGAAACAAAAGCCCACCGUCAACACCCCCAAUCCGCCGCAUUCGCCACAUUUUGGCCAGGUUU--UUGGCCACCAGUGUUGACACAA .((((................(((........)))........))))...((((((((............((....)).......(((((((....--)))))))...)))))))).... ( -27.95) >DroSim_CAF1 6101 118 - 1 GGGGCACACAAAAACACACAACUCAAAAGGCCGAGAAACAAAAGCCCACCUUCAACACCCCCAAUCCGCCGCAUUCGCCACAUUUUGGCCAGGUUU--UUGGCCACAAGUGUUGACACAA .((((................(((........)))........))))....(((((((............((....)).......(((((((....--)))))))...)))))))..... ( -23.95) >DroEre_CAF1 160 114 - 1 GGGGCACACAAAAACACACAACUCAAAA-GCCGAGAAACAA-AGCCUACCUUCAACACCCCCCAUCCGCAACAUUCGCC--UUUUUGGCCAGGUUU--UUGGCUACAAGUGUUGACACAA ((((.................(((....-...))).....(-((.....))).......))))....((((((((.(..--....(((((((....--)))))))).))))))).).... ( -19.20) >DroYak_CAF1 842 115 - 1 GGGGCACA-AAAAACACACAACUCAAAA-GCCGAGAAACAA-AGCCUACCUUCAACACCACCCAUCAGCAAGAUUCGCC--UUUUUGGCCAGGUUUUUUUGGCCACAAGUGUUGACACAA .((((...-............(((....-...)))......-.))))....(((((((.........((.......)).--....((((((((....))))))))...)))))))..... ( -22.65) >consensus GGGGCACACAAAAACACACAACUCAAAAGGCCGAGAAACAAAAGCCCACCUUCAACACCCCCAAUCCGCCGCAUUCGCCACAUUUUGGCCAGGUUU__UUGGCCACAAGUGUUGACACAA .((((................(((........)))........))))....(((((((.........((.......)).......(((((((......)))))))...)))))))..... (-21.29 = -21.09 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:01 2006