
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,466,960 – 16,467,080 |
| Length | 120 |
| Max. P | 0.654445 |

| Location | 16,466,960 – 16,467,080 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -17.59 |
| Energy contribution | -18.26 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16466960 120 + 22224390 UUUGAUUGUUCAACUGGUUCCUCACCGCUUGGUCGUCAUGCAAAUCGCUCGCAUAAACAAACAAACAAAUAACGAACAAAUCAAAUAAAAUGAAACUGGGAAAACGAGGAGGAACUAUGA .((((....)))).(((((((((..((((..((..((((((.....)).........................((.....)).......)))).))..))....))..)))))))))... ( -23.30) >DroSec_CAF1 5099 92 + 1 UUUGAUUGUUCAACUGGUUCCUCACCGUUUGGUCGUCAUGCAAGU----------------------------AAACAAAUCAAAUAAAAUGAAACUGGGAAAACGAGGAGGAACUAUGA .((((....)))).(((((((((..(((((..(((((((......----------------------------................))))...)))..)))))..)))))))))... ( -22.45) >DroSim_CAF1 1960 92 + 1 UUUGAUUGUUCAACUGGUUCCUCACCGUUUGGUCGUCAUGCAAGU----------------------------AAACAAAUCAAAUAAAAUGAAACUGGGAAAACGAGAAGGAACUAUGA .((((....)))).((((((((...(((((..(((((((......----------------------------................))))...)))..)))))...))))))))... ( -16.35) >consensus UUUGAUUGUUCAACUGGUUCCUCACCGUUUGGUCGUCAUGCAAGU____________________________AAACAAAUCAAAUAAAAUGAAACUGGGAAAACGAGGAGGAACUAUGA .((((....)))).(((((((((..(((((..(((((((..................................................))))...)))..)))))..)))))))))... (-17.59 = -18.26 + 0.67)

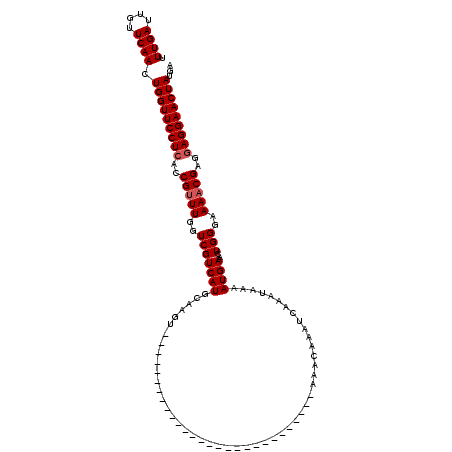
| Location | 16,466,960 – 16,467,080 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -16.10 |
| Energy contribution | -15.66 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16466960 120 - 22224390 UCAUAGUUCCUCCUCGUUUUCCCAGUUUCAUUUUAUUUGAUUUGUUCGUUAUUUGUUUGUUUGUUUAUGCGAGCGAUUUGCAUGACGACCAAGCGGUGAGGAACCAGUUGAACAAUCAAA (((..(((((((.((((..........(((.......))).(((.(((((((....((((((((....)))))))).....))))))).))))))).)))))))....)))......... ( -31.10) >DroSec_CAF1 5099 92 - 1 UCAUAGUUCCUCCUCGUUUUCCCAGUUUCAUUUUAUUUGAUUUGUUU----------------------------ACUUGCAUGACGACCAAACGGUGAGGAACCAGUUGAACAAUCAAA (((..(((((((.((((((.....((((((.......)))..(((..----------------------------....))).)))....)))))).)))))))....)))......... ( -18.40) >DroSim_CAF1 1960 92 - 1 UCAUAGUUCCUUCUCGUUUUCCCAGUUUCAUUUUAUUUGAUUUGUUU----------------------------ACUUGCAUGACGACCAAACGGUGAGGAACCAGUUGAACAAUCAAA (((..(((((((..(((((.....((((((.......)))..(((..----------------------------....))).)))....)))))..)))))))....)))......... ( -16.80) >consensus UCAUAGUUCCUCCUCGUUUUCCCAGUUUCAUUUUAUUUGAUUUGUUU____________________________ACUUGCAUGACGACCAAACGGUGAGGAACCAGUUGAACAAUCAAA (((..(((((((.((((((........(((.......))).(((((.....................................)))))..)))))).)))))))....)))......... (-16.10 = -15.66 + -0.44)

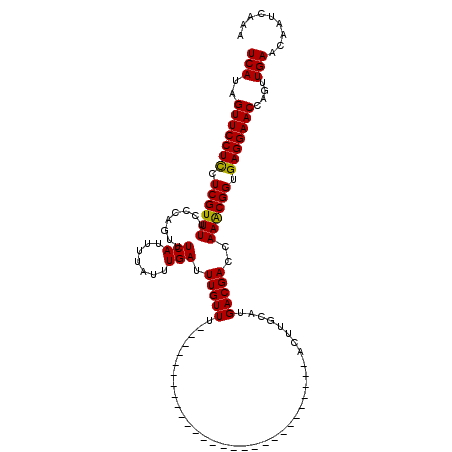
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:54 2006