
| Sequence ID | X_DroMel_CAF1 |
|---|---|
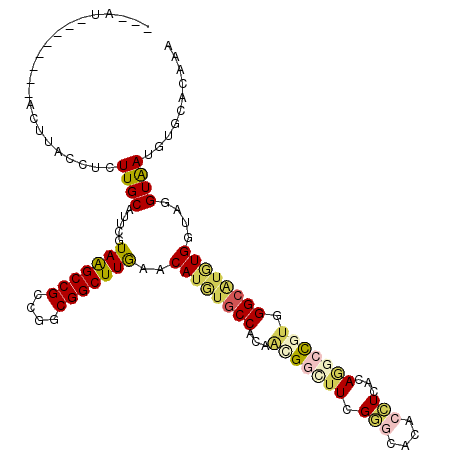
| Location | 16,460,782 – 16,460,929 |
| Length | 147 |
| Max. P | 0.897189 |

| Location | 16,460,782 – 16,460,895 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -38.25 |
| Consensus MFE | -25.75 |
| Energy contribution | -26.45 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16460782 113 + 22224390 ---GUGG-CUGCUACUGACCUCUUGCAUUCGUAAGCCGCCGGCGGCUUGAACAUGUGCCACAAAGGCUUCGGGCACACCUCGCAAGCCGUGGGCAUGUGGUAGGUAAUGUGAACGAA ---.((.-(((((((......(((((....)))))..(((.((((((((....((((((............)))))).....)))))))).)))..))))))).))........... ( -44.60) >DroVir_CAF1 17432 112 + 1 ---CUG--CAGUUGCUUACCUCUUGCAUUCGUAGGCCGCCGGCGGCUUGAACAUGGACCACAGCGGCUUGGCGCAAACGUCACAGAACGAGGGCGUAUGAUACGUAAUGUGCACGAA ---.((--(((((((.((.(...(((.((((((((((((.(..((((.......)).)).).)))))))((((....)))).....))))).)))...).)).))))).)))).... ( -33.40) >DroPse_CAF1 297 107 + 1 ---A-------GCACAAACCUCUUGCAUUCGUAAGCCGCCGGCGGCUUGAACAUGUGCCACAAUGGCUUCGGGCACACCUCGCACGCUGUGGGCAUGUGGUAGGUGAUGUGCACAAA ---.-------(((((.((((...((((...(((((((....)))))))...))))((((((.((.(....).))...(((((.....)))))..))))))))))..)))))..... ( -42.40) >DroGri_CAF1 302 104 + 1 -------------ACUUACCUCUUGCAUUCGUAGGCCGCCGGCGGCUUAAACAGUUGCCACAGCGGCUUGGAGCAUAAAUCACAGAACGUUGGCGUAUGGUACGUAAUGUGCACAAA -------------((.((((...(((...((((((((((.(((((((.....)))))))...))))))).((.......)).....)))...)))...)))).))............ ( -31.80) >DroWil_CAF1 283 117 + 1 UUGAUGGCAUCCCACUUACCUUUUGCAUUCAUAAGCCGCCGGCGGCUUGAACAUAUGCCACAAGGGUUUCGGACACACCUCACAAGCUGUGGGCAUGUGAUAGGUAAUAUGAACAAA (((.(((....))).((((((..........(((((((....)))))))..((((((((.((..(((((..((......))..))))).))))))))))..))))))......))). ( -33.40) >DroAna_CAF1 290 102 + 1 ---GU------------ACCUCUUGCAUUCGUAAGCCGCCGGCGGCUUGAACAUGUGCCACAACGGCUUCGGGCACACCUCGCAGGCCGUGGGCAUGUGGUAGGUGAUAUGGACGAA ---((------------.((.(((((.....(((((((....)))))))..((((((((...(((((((((((.....)))).))))))).)))))))))))))......))))... ( -43.90) >consensus ___AU________ACUUACCUCUUGCAUUCGUAAGCCGCCGGCGGCUUGAACAUGUGCCACAACGGCUUCGGGCACACCUCACAGGCCGUGGGCAUGUGGUAGGUAAUGUGCACAAA ......................((((.....(((((((....)))))))..((((((((...(((((((.(((....)))...))))))).))))))))....)))).......... (-25.75 = -26.45 + 0.70)


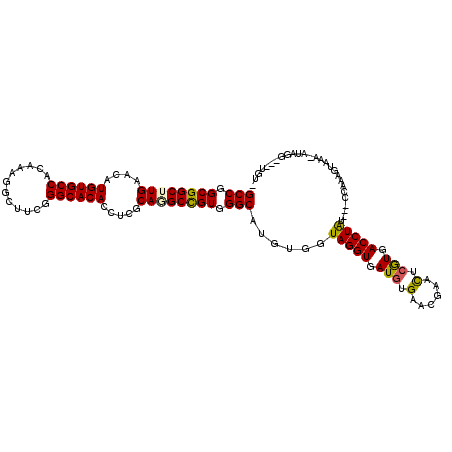
| Location | 16,460,815 – 16,460,929 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -41.12 |
| Consensus MFE | -36.85 |
| Energy contribution | -36.47 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16460815 114 + 22224390 GCCGGCGGCUUGAACAUGUGCCACAAAGGCUUCGGGCACACCUCGCAAGCCGUGGGCAUGUGGUAGGUAAUGUGAACGAACUCGUGACCUAU---CCAAAACAAAAAAGGGUGUUGU- (((.((((((((....((((((............)))))).....)))))))).))).((.(((((((.(((.(......).))).))))))---))).((((........))))..- ( -42.90) >DroPse_CAF1 324 106 + 1 GCCGGCGGCUUGAACAUGUGCCACAAUGGCUUCGGGCACACCUCGCACGCUGUGGGCAUGUGGUAGGUGAUGUGCACAAAUUCAUGACCUA-----CAAAGUAAA--CAG----UUA- (((.(((((.((....((((((............)))))).....)).))))).))).....((((((.(((..........))).)))))-----)........--...----...- ( -36.50) >DroEre_CAF1 328 113 + 1 GCCGGCGGCUUGAACAUGUGCCACAAAGGCUUCGGGCACACCUCACAGGCCGUGGGCAUGUGGUAGGUGAUGUGGACGAACUCGUGACCUAU---CCAGAUGA-A-AUGGGUUUAGUG (((.((((((((....((((((............)))))).....)))))))).)))((.((((((((.(((.(......).))).))))).---))).))..-.-............ ( -45.20) >DroYak_CAF1 309 112 + 1 GCCGGCGGCUUGAACAUGUGCCACAAAGGCUUCGGGCACACCUCGCAGGCCGUGGGCAUGUGGUAGGUGAUGUGAACGAACUCGUGACCUAU---CCAAAUCA-A-AUAAGUUUAGU- (((.((((((((....((((((............)))))).....)))))))).))).((.(((((((.(((.(......).))).))))))---))).....-.-...........- ( -44.30) >DroAna_CAF1 312 109 + 1 GCCGGCGGCUUGAACAUGUGCCACAACGGCUUCGGGCACACCUCGCAGGCCGUGGGCAUGUGGUAGGUGAUAUGGACGAACUCGUGACCUGGAAGCGCAAGU-AG-AUGAG------- ((..(((.(((...((((((((...(((((((((((.....)))).))))))).)))))))).(((((.((..(......)..)).))))).))))))..))-..-.....------- ( -41.30) >DroPer_CAF1 324 106 + 1 GCCGGCGGCUUGAACAUGUGCCACAAUGGCUUCGGGCACACCUCGCACGCUGUGGGCAUGUGGUAGGUGAUGUGCACAAAUUCAUGACCUA-----CAAAGUAAA--CAG----UAA- (((.(((((.((....((((((............)))))).....)).))))).))).....((((((.(((..........))).)))))-----)........--...----...- ( -36.50) >consensus GCCGGCGGCUUGAACAUGUGCCACAAAGGCUUCGGGCACACCUCGCAGGCCGUGGGCAUGUGGUAGGUGAUGUGAACGAACUCGUGACCUAU___CCAAAGUAAA_AUAGG___UGU_ (((.((((((((....((((((............)))))).....)))))))).)))......(((((.(((.(......).))).)))))........................... (-36.85 = -36.47 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:48 2006